Abstract
Lycodes brevipes is one of the representative species of zoarcid fishes with little genetic information. The complete mitochondrial genome sequence of L. brevipes was sequenced by long PCR and primer walking methods. The mitogenome is a circular molecule of 16,537 bp in length including the structure of 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes and two non-coding regions (origin of light strand replication and control region). The present study also revealed the phylogenetic relationship of this species at the molecular level using NJ algorithm. The mitogenome of L. brevipes provides essential genetic data for further phylogenetic and evolutionary analyses.
The Chukchi Sea is located at high latitudes and one of the most sensitive waters to climate change. In polar marine ecosystem, the environmental characteristics seriously shape the temporal and spatial distributions of fishes. The eelpouts of genus Lycodes is comprised of about 62 species as the largest group of zoarcid fishes distributed in the Arctic Ocean and adjacent waters, and is well known for currently the most species-rich genus, providing a great potential for Arctic marine speciation and genetic analysis (Anderson Citation1994; Møller and Gravlund Citation2003). However, species identification of Lycodes is often difficult due to similar morphological characteristics.
Lycodes brevipes Bean, 1890 is one of the representative species of this genus with little phylogenetic analysis (Møller and Gravlund Citation2003). To understand its genetic background and clarify the phylogenetic position, sufficient genetic information of L. brevipes is needed and helpful for effective management and conservation of this species.
The voucher specimen was collected from Chukchi Sea continental shelf (168.2°W, 69.3°N) during the 6th Chinese National Arctic Research Expedition (CHINARE 6) in 2014, and deposited at the Third Institute of Oceanography, SOA. In this study, the complete mitochondrial genome (mitogenome) of L. brevipes was sequenced and phylogenetic topology of Lycodes species was reconstructed.
Long PCR and primer walking methods are employed to amplify the mitogenome sequence of L. brevipes with 37 pairs of primers (Miya and Nishida Citation1999). It was a typical circular form with 16,537 bp in length (GenBank number MK133251) and comprised of 37 coding and two non-coding regions. The 37 coding regions include 13 protein-coding genes, 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12S and 16S rRNAs), and two mainly non-coding regions (origin of light strand replication/OL and control region/CR). Most of these genes are encoded on the H-strand, except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) that are oriented to the L-strand. The arrangement of all genes is in accordance with most other vertebrates (Cheng et al. Citation2012; Wang et al. Citation2018). The overall base composition is as follows: 25.6% for A, 25.0% for T, 18.6% for G, and 30.8% for C, with a slight A + T-rich feature (50.6%).
Adjacent genes overlap by 31 bp in nine different locations from 1 to 10 bp, smaller than those in most mammals (40–46 bp; Broughton et al. Citation2001), and have gaps of 43 bp in nine different locations from 1 to 24 bp. The putative OL is 40 bp long, located in the WANCY region (tRNATrp-tRNAAla-tRNAAsn-tRNACys-tRNATyr). The CR of L. brevipes located between the tRNAPro and the tRNAPhe genes is determined to be 865 bp in length. Except for COI starting with GTG, the remaining 12 protein-coding genes start with ATG. Seven protein-coding genes are inferred to terminate with TAA (ND1, ND2, COI, ATP8, ATP6, COIII, and ND4L), three with TAG (ND3, ND5, and ND6), and three with incomplete stop codons T-- (COII, ND4, and Cyt b).
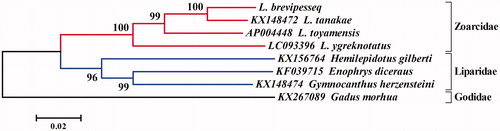
Phylogenetic relationship was reconstructed by MEGA 6.06 (Tamura et al. Citation2013) based on the mitogenome sequences of L. brevipes and seven other Arctic fish species. The NJ tree () also showed that all species can be easily distinguished from each other with the high bootstrap values. The species L. brevipes was closer to the generic species than other species. The genus Lycodes species first formed a monophyly, and then they clustered with other Arctic fish species. The complete mitochondrial genome of L. brevipes reported in this study would facilitate further phylogenetic analysis and population genetics of this species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson ME. 1994. Systematics and osteology of the Zoarcidae (Teleostei: Perciformes). J.L.B. Smith Institute of Ichthyology. Ichthyol Bull. 60:1–120.
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Cheng J, Ma GQ, Song N, Gao TX. 2012. Complete mitochondrial genome sequence of bighead croaker Collichthys niveatus (Perciformes, Sciaenidae): a mitogenomic perspective on the phylogenetic relationships of Pseudosciaeniae. Gene. 491:210–223.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei: Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426.
- Møller PR, Gravlund P. 2003. Phylogeny of the eelpout genus Lycodes (Pisces, Zoarcidae) as inferred from mitochondrial cytochrome b and 12S rDNA. Mol Phylogenet Evol. 26:369–388.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Wang SQ, Zhao LL, Li Y, Zhang ZH, Wang ZL, Gao TX. 2018. Complete mitochondrial DNA genome of grey pomfret Pampus cinereus (Bloch, 1795) (Perciformes: Stromateidae). Mitochondrial DNA B Resour. 3:683–684.