Abstract
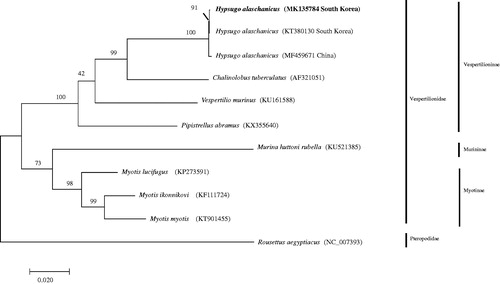
The mitogenome of the Hypsugo alaschanicus is a circular module of 16,911 bp, which consists of 37 genes, containing 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes (12S rRNA and 16S rRNA), and a control region. The mitogenome of H. alaschanicus is AT-biased, with a nucleotide composition of 33.9% A, 30.8% T, 22.5% C, and 12.8% G. The phylogenetic analysis revealed that two Korean H. alaschanicus individuals were well grouped and formed a sister to the Chinese individual.
Hypsugo alaschanicus (Vespertilionidae) is a small bat species, found widely in East Palearctic region of Mongolia, China, east part of Russia, Korea, and Japan (Horáček et al. Citation2000; Jo et al. Citation2012). Horácek et al. (Citation2000) suggested that South Korean H. alaschanicus might represent a separate subspecies as H. a. coreensis, but Yoshiyuki (Citation1989) treated the Korean H. alaschanicus as a distinct species (Pipistrellus coreensis imaizumi). A wing tissue for DNA extraction was collected from an individual of H. alaschanicus in Hwacheon-gun (N38 04 11.1 E127 46 42.0). After collecting wing tissue samples by punching, the captured individual was released. The voucher specimen (KNUWB-2) was deposited in the Wildlife and Fish Conservation Center of the Institute of Forest Science, Kangwon National University. A previously published mitogenome (KT380130) of H. alaschanicus (Kim and Park Citation2015) was used as a reference for designing PCR primers, and gene annotation using Geneious 11.1.5 (Kearse et al. Citation2012). Phylogenetic tree was constructed using maximum-likelihood (ML) method procedures implemented in MEGA-X (Kumar et al. Citation2018). The Egyptian fruit bat Rousettus aegyptiacus (NC_007393) was used as an outgroup.
The complete mitogenome (MK135784) of H. alaschanicus contains 16,911 bp in length, which consists of two non-coding regions (replication origin (OL) and control region) and conserved set of 37 vertebrate mitochondrial genes including 13 protein-coding genes, 2 rRNA genes (12S rRNA and 16S rRNA) and 22 tRNA genes. The order and orientation of gene arrangement are similar to those of other vertebrates (Kim and Park Citation2012; Kim et al. Citation2013; Yoon et al. Citation2013; Kim et al. Citation2017; Lim et al. Citation2017; Lim et al. Citation2018).
The mitogenome of H. alaschanicus is AT-biased, with a nucleotide composition of 33.9% A, 30.8% T, 22.5% C, 12.8% G. Length of the two rRNA genes (12s rRNA and 16s rRNA) were 955 bp (60.7% A + T) and 1563 bp (64.1% A + T), respectively.
Total length of the 13 mitochondrial protein-coding genes of H. alaschanicus is 11,376 bp long (64.9% A + T) which encodes 3792 amino acids except stop codons. Mitochondrial PCGs of H. alaschanicus use start codon ATG, except ND2, ND3 (ATT) and ND5 (ATA). Stop codon TAA is used in eight PCGs (ND1, Cox1, Cox2, Atp8, Atp6, Nd4L, Nd5, and Nd6 except for Cytb (AGA)). Total 22 tRNA genes, including two leucine-tRNA genes (tRNALeu(UUR) and tRNALeu(CUN)) and two serine-tRNA genes (tRNASer(AGY) and tRNASer(UCN)), were present in the mitogenome. The OL region is 36 bp long and located between tRNAAsn and tRNACys within the WANCY region containing the five tRNA genes (tRNATrp, tRNAAla, tRNAAsn, tRNACys, and tRNATyr). The D-loop region is 1480 bp in length, with A + T content of 64.8% and located between tRNAPro and tRNAPhe.
Except for D-loop, the mitogenome of H. alaschanicus (MK135784) has 99.9% sequence similarity to that of the other Korean H. alaschanicus (KT380130), whereas 99.6% sequence similarity to the Chinese H. alaschanicus (MF459671).
The phylogenetic analysis revealed that two Korean H. alaschanicus individuals (MK135784 and KT380130) were well grouped and formed a sister to the Chinese individual (MF459671). Hypsugo alaschanicus formed a sister clade to Chalinolobus tuberculatus (AF321051) ().
Figure 1. The phylogenetic relationship of Hypsugo alaschanicus based on mitogenome sequences. The phylogenetic tree was generated using the maximum-likelihood method based on GTR + G + I model. The robustness of the tree was tested with 2000 bootstraps. The numbers on the branches indicate bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Horáček I, Vladimír H, Jiří G. 2000. Bats of the Palearctic region: a taxonomic and biogeographic review. Proceedings of the VIIIth European bat research symposium. Kraków: CIC ISEZ PAN.
- Jo YS, Kim TW, Choi BJ, Oh HS. 2012. Current status of terrestrial mammals on Jeju Island. J Species Res. 1:249–256.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim HR, Jeon MG, Min JH, Kim HJ, Park YC. 2017. Complete mitochondrial genome of the roe deer Capreolus pygargus tianschanicus (Cervidae) from Korea. Mitochondrial DNA B Resour. 2:558–559.
- Kim EK, Kim HR, Jeon SH, Park YC. 2013. Complete mitochondrial genome of a water deer subspecies, Hydropotes inermis argyropus (Cervidae: Hydropotinae). Mitochondrial DNA. 24:17–18.
- Kim HR, Park YC. 2012. The complete mitochondrial genome of the Korean red-backed vole, Myodes regulus (Rodentia, Murinae) from Korea. Mitochondrial DNA. 23:148–150.
- Kim JY, Park YC. 2015. Gene organization and characterization of the complete mitogenome of Hypsugo alaschanicus (Chiroptera: Vespertilionidae). Genet Mol Res. 14:16325–16331.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lim SJ, Kim HR, Cho JY, Park YC. 2017. Complete mitochondrial genome of the least weasel Mustela nivalis (Mustelidae) in Korea. Mitochondrial DNA B Resour. 2:740–741.
- Lim SJ, Kim KY, Kim HR, Chae HM, Park YC. 2018. Phylogenetic position of the Siberian flying squirrel Pteromys volans based on complete mitochondrial genome sequences. Mitochondrial DNA B Resour. 3:601–602.
- Yoon KB, Kim JY, Kim HR, Cho JY, Park YC. 2013. The complete mitochondrial DNA genome of a greater horseshoe bat subspecies, Rhinolophus ferrumequinum quelpartis (Chiroptera: Rhinolophidae). Mitochondrial DNA. 24:22–24.
- Yoshiyuki M. 1989. A systematic study of the Japanese Chiroptera. National Science Museum Monographs. 7:1–242.
