Abstract
Siphonostegia chinensis Benth. is a common Chinese herbal medicine widely distributed in eastern Asia. The complete chloroplast genome of S. chinensis was newly sequenced in this study. The total chloroplast genome size was 148,961 bp, its structure and gene contents were well conserved as typical chloroplast characteristics. The overall guanine-cytosine (GC) content of the S. chinensis chloroplast genome is 36.4%. The chloroplast genome encodes 130 unique genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. We used the cp genome of S. chinensis and 20 other chloroplast genomes to perform a phylogenetic analysis, which indicated that S. chinensis is sister to Schwalbea americana in Orobanchaceae.
The genus Siphonostegia of the family Orobanchaceae includes three species and two are distributed in China and also widely distributes in Japan, North Korea, and Russia (Mcneal et al. Citation2013). Siphonostegia chinensis is an annual herb with yellow and red-purple slender corolla tube. It was traditionally used to treat icterohepatitis, dysuria, and external bleeding. Recent study showed it contains acteoside, isoacteoside, and crenatoside, and its extracts could be potentially used as new natural active ingredients for whitening cosmetics (Wang Citation2008; Kim et al. Citation2009). Here, we assembled and characterized the first complete chloroplast genome of S. chinensis, and analysed genetic relationships with the closely related species in Orobanchaceae.
Experimental samples were collected from Chunan, Hangzhou, China (Voucher No. ZSTU00896, deposited at Zhejiang Sci-Tech University). Total DNA was extracted from silica-dried leaves of S. chinensis individual using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). The complete chloroplast genome of platform sequences was generated using Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, ca. 32.8 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. Then, CLC Genomics Workbench (CLC Bio, Aarhus, Denmark) was used for trimming, de novo assembly, and mapping to Rehmannia chingii (KX426347) as reference. BLAST, GeSeq (Tillich et al. Citation2017), and Geneious version 11.1.2 (Biomatters Ltd, Auckland, New Zealand) were used to align, assemble, and annotate the plastome.
The total chloroplast genome size of S. chinensis (GenBank Accession No. MK113828) was 148,961 bp. The total length is composed of an LSC (84,124 bp), two IRs (49,592 bp), and an SSC (15,245 bp). The overall GC content of the S. chinensis chloroplast genome is 36.4%. The chloroplast genome encodes 130 unique genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The tRNA genes are distributed throughout the whole genome with 22 in the LSC, 1 in the SSC, 14 in the IR regions, while rRNAs only situate in IR regions. Seventeen genes had two copies, which included six PCG genes (rpl2, rpl23, ycf2, ycf15, ndhB, and rps7), seven tRNA genes (tRNA-CAU, tRNA-CAA, tRNA-GAC, tRNA-UGC, tRNA-ACG, and tRNA-GUU), and four all rRNA genes (rrn16, rrn23, rrn4.5, and rrn5). Among the protein-coding genes, one gene (ycf3) contained two introns, and six genes (rps16, petB, rpl16, rpl2, ndhB, and ndhA) had one intron each.
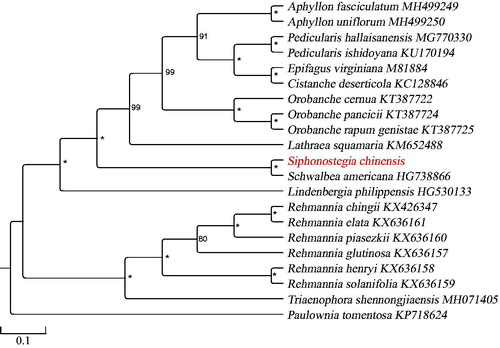
Twenty-one chloroplast genomes sequences from Orobanchaceae and Paulowniaceae were aligned with MAFFT version 7.3 (Katoh and Standley Citation2013), and the maximum likelihood (ML) inference was performed using TVM + F+R3 model with 1000 bootstrap replicates with IQ-TREE version.1.6.7 (Nguyen et al. Citation2015). The TreeGraph2 version.2.14.0 (Müller and Stöver Citation2010) were used to edit and modify tree file. The result revealed that S. chinensis is sister to Schwalbea americana in Orobanchaceae ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Chou G, Zhu E, Lee SY, Kim YH, Park SK, Oh ST, Kim KH. 2009. The study on the whitening effects of traditional Chinese medicines. J Soc Cosmet Sci Korea. 35:257–263.
- Mcneal JR, Bennett JR, Wolfe AD, Sarah M. 2013. Phylogeny and origins of holoparasitism in Orobanchaceae. Am J Bot. 100:971–983.
- Müller KF, Stöver BC. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 11:7–7.
- Nguyen LT, Schmidt HA, Von HA, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Wang JL. 2008. Research progress of chemical constituents and pharmacological effects of Siphonostegia chinensis Benth. Heilongjiang Med. 31:61–62.