Abstract
Sanicula chinensis Bunge is a widespread and important Chinese medicinal plant. In this study, we reported the complete chloroplast genome of Sanicula chinensis. The genome sequence was 154,650 bp in length, including a large single copy region (LSC) of 85,263 bp and a small single copy region (SSC) of 16,927 bp, which were separated by two inverted repeat (IR) regions of 26,230 bp. The complete chloroplast genome contains 129 genes, including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis with several reported chloroplast genomes (Apioideae and Hydrocotyloideae) showed that S. chinensis has a close genetic relationship with Apioideae.
Sanicula chinensis Bunge is a typical East Asian distribution and perennial herb. As an important Chinese medicinal plant, it is reported that it is effective in the treatment of rheumatalgia, cough, activating blood circulation (Zhou and Jia Citation2007). Furthermore, based on previous phylogenetic analyses of molecular data, Sanicula is not monophyletic(Downie et al. Citation2000; Calvino and Downie Citation2007). Considering that chloroplast DNA (cpDNA) conservativity and its slow rate of nucleotide substitution make cpDNA widely used in plant phylogeny (Olmstead and Palmer Citation1994); thus, we first report the complete chloroplast genome of S. chinensis, which will help in molecular and phylogenetical studies of this plant.
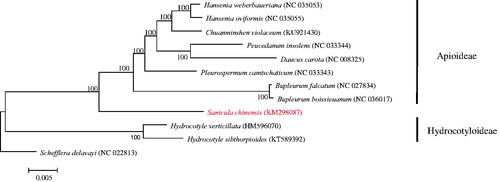
Fresh leaves of S. chinensis were collected from Shennongjia Scenic Area (31°15 N, 109°56 E), Hubei Province, China. Voucher specimens were deposited in Hiaoqiao University Herbarium (00007622). Total genomic DNA was extracted by Plant Genomic DNA Kit (Sangon Biotech, Shanghai, China). Paired-end reads were sequenced by using Illumina Hiseq Platform (Illumina, San Diego, CA). Approximately 10 Gb of paired-end (150 bp) sequence data were randomly extracted from the total sequencing output and used as input for NOVOPlasty (Dierckxsens et al. Citation2017) to assemble the plastid genome. The plastid genome of Pleurospermum camtschaticum (GenBank accession number: NC_033343) was used as the seed sequence. The genes in chloroplast genome were predicted using Geneious version 11.0.4 (Kearse et al. Citation2012) and corrected manually. The phylogeny based on the complete plastid genome shared by Sanicula species was inferred from the ML tree using MEGA7.0 (Kumar et al. Citation2016) from alignments created by the MAFFT (Katoh et al. Citation2002) using plastid genomes of 12 species.
The complete chloroplast genome of S.chinensis (GenBank accession number: MK208987) was 154,650 bp in total sequence length with 38.20% GC contents. Four distinct regions were separated by the complete chloroplast, such as large single copy (LSC) region was 85,263 bp, small single copy (SSC) region was 16,927 bp, and a pair of inverted repeat regions are 26,230 bp in each length. The chloroplast genome detected a total of 129 genes including 84 protein-coding genes, 37 tRNA genes, and 8 rRNA genes.
The phylogenetic analysis of 12 chloroplast genomes showed that S.chinensis is closely related to Apioideae (). This complete cp genome can be further used for population genomic studies, phylogenetic analyses, genetic engineering studies of Sanicula. Such genomic and genetic information would be fundamental to medicinal research.
Acknowledgments
The authors thank Professor Yan Yu (College of Life Sciences, Sichuan University) for the help of sequence analysis.
Disclosure statement
The authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Calvino CI, Downie SR. 2007. Circumscription and phylogeny of Apiaceae subfamily Saniculoideae based on chloroplast DNA sequences. Mol Phylogenet Evol. 44:175–191.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Downie SR, Katz-Downie DS, Watson MF. 2000. A phylogeny of the flowering plant family Apiaceae based on chloroplast DNA rpl16 and rpoC1 intron sequences: towards a suprageneric classification of Subfamily Apioideae. Am J Bot. 87:273–292.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol Evol. 33:1870.
- Olmstead RG, Palmer JD. 1994. Chloroplast DNA systematics: a review of methods and data analysis. Am J Bot. 81:1205–1224.
- Zhou XJ, Jia MR. 2007. Investigation on the medicinal resources of folk ethnic medicine Sanicula. Chin J Ethnomed Ethnopharm. 89:366–367.