Abstract
Goodyera schlechtendaliana is a common orchid species in East Asia. A distinctive population of G. schlechtendaliana with the lateral appendages of the column in their flowers was found in southwestern Korea. In this study, we presented complete chloroplast genome of this morphotype as a part of systematic study of the Goodyera. The chloroplast genome is 153,882 bp in length and contains 134 genes (83 CDSs, 8 rRNAs, and 39 tRNAs). Phylogenetic analysis showed that the morphotype of G. schlechtendaliana with column appendages is sister to a normal form of G. schlechtendaliana with long branch, supporting that this distinctive morphotype has a potential to be a new species.
Goodyera schlechtendaliana Rchb. f. (Orchidaceae: Orchidoideae) is a common orchid widely distributed in East Asia ranging from the Himalayas, Sumatra, China, Taiwan, Korea, to Japan (Chen et al. Citation2009). It is characterized by having white variegated marking on the adaxial surface of the leaves that are arranged in basal rosette in Orchidaceae. We found a population of plants similar to G. schlechtendaliana but having distinct morphology that would be potentially a new species on remote islands in southwestern Korea. This plant is different from G. schlechtendaliana in having larger leaves, longer inflorescence, and the lateral appendages of the column. As part of the systematic study to understand the phylogenetic position of this morphotype in Goodyera we determined its complete chloroplast genome.
A plant used in this study was collected on Hongdo Island in Heuksan-myeon, Shinan-gun, Jeollanam-do province, Korea (voucher specimen: Oh 7169, deposited in Daejeon University Herbarium, TUT). Total genomic DNA was extracted from fresh leaves using a DNeasy Plant Mini kit (QIAGEN, Hilden, Germany). Paired-end sequencing was performed using HiSeq4000 (Illumina, San Diego, CA) and de novo assembly was performed using Velvet version 1.2.10 (Zerbino and Birney Citation2008) and gap sequences were filled by SOAPGapCloser version 1.12 (Zhao et al. Citation2011). Geneious R11 version 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for genome annotation.
The chloroplast genome of the morphotype of G. schlechtendaliana with the column appendages (GenBank accession number, MK134679) is 153,882 bp in length and composed of four subregions: 82,733 bp of large single copy (LSC), 18,019 bp of small single copy (SSC) regions, and 26,535 bp of a pair of inverted repeats (IRs). It contains 134 genes (83 CDS, 8 rRNAs, and 39 tRNAs); 20 genes (eight CDS, four rRNAs, and eight tRNAs) are in each IR region. The overall GC content of the morphotype of G. schlechtendaliana was 37.2% and those in the LSC, SSC, and IR regions were 34.9, 29.7 and 43.3%, respectively.
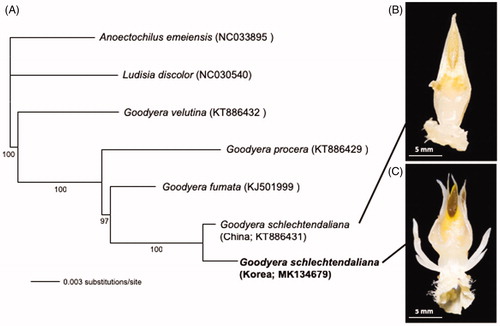
Complete chloroplast genomes from five species of Goodyera and two outgroups, Anoectochilus emeiensis (NC033895) and Ludisia discolor (NC030540), were included in phylogenetic analysis using the maximum likelihood method. Sequence alignment was conducted by MAFFT version 7.388 (Katoh and Standley Citation2013), and phylogenetic tree was constructed by IQ-TREE version 1.6.6 (Nguyen et al. Citation2014) with 1000 pseudoreplicates of bootstrapping.
The phylogenetic tree shows that the morphotype of G. schlechtendaliana with column appendages from Korea is sister to G. schlechtendaliana with 100% bootstrap support. A long branch leading to the morphotype of G. schlechtendaliana with column appendages suggests that there is large amount of evolutionary divergence between the two samples (). The chloroplast genome data are consistent with morphology, supporting that the morphotype of G. schlechtendaliana from Korea could potentially be treated as a new species. In addition, the chloroplast genome will be a useful resource for understanding phylogenetic relationship of the genus Goodyera.
Figure 1. A maximum-likelihood tree (A) using chloroplast genomes of the morphotype of G. schlechtendaliana with the column appendages from Korea (this study) and previously published related taxa: G. schlechtendaliana from China (KT886431), Goodyera fumata (KJ501999), Goodyera procera (KT886429), Goodyera velutina (KT886432), Ludisia discolor (NC030540), and Anoectochilus emeiensis (NC033895). The number below the branch indicates bootstrap value. The columns of G. schlechtendaliana (B) and the morphotype of G. schlechtendaliana with lateral appendages (C) are shown.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen X, Lang K, Gale S, Cribb P, Ormerod P. 2009. Goodyera. Flora of China. Vol. 25. Beijing: Science Press; p.45–54.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
