Abstract
Ixonanthes chinensis (Ixonanthaceae) is an endangered tree species distributed in South China and Vietnam. The phylogenetic position of Ixonanthes in the order Malpighiales remains controversial. Here we reported and characterized the complete chloroplast genome sequence of I. chinensis assembled from Illumina sequencing reads. The chloroplast genome size is 155,296 bp, containing a large single copy (LSC) region of 83,829 bp, a short single copy (SSC) region of 18,917 bp, and a pair of inverted repeats (IRs) of 26,275 bp. We predicted a total of 139 genes, which included 87 protein-coding genes, 44 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that I. chinensis was sister to Linum usitatissimum from the family Linaceae rather than Erythroxylum novogranatense from the family Erythroxylaceae.
Ixonanthes chinensis Champ., a tree species in the family Ixonanthaceae, has been widely distributed in tropical and subtropical evergreen forest in South China. Due to the excessive timber harvest and habitat destruction, however, the population of I. chinensis has decreased rapidly and become endangered in China (Fu Citation1992; Wu Citation2000). It was considered as ‘Vulnerable’ category in the IUCN Red List (Sun Citation1998). Ixonanthes and Erythroxylum were traditionally placed in the family Erythroxylaceae, but recent molecular phylogenetic studies indicated that they belonged to two distinct families, with Ixonanthes in the family Ixonanthaceae and Erythroxylum in the family Erythroxylaceae (Xi et al. Citation2012). The complete chloroplast genome sequence of I. chinensis will provide useful information for both its phylogenetic resolution and conservation genetics. In this study, we obtained the complete chloroplast genome of I. chinensis using Illumina sequencing technology, and the sequence and genome annotation are available in GenBank under accession number MK163924.
DNA extraction was performed using the silica-gel dried leaves of a single individual of I. chinensis collected from Dapeng, Shenzhen, Guangdong, China. A shotgun DNA library was constructed with an insert size of 350 bp and then sequenced on an Illumina Hiseq X Ten platform. 10.7 Gb paired-end reads (PE = 150 bp) were generated and then used as an input into NOVOPlasty (Dierckxsens et al. Citation2017) to assemble the chloroplast genome using rbcL gene sequence of the same species (GenBank accession number AB233893) as the seed sequence. The DOGMA online tool (Wyman et al. Citation2004) was used to annotate the chloroplast genome, and further manual adjustment was conducted when necessary. The gene map of this species was generated by the online program OGDRAW (Lohse et al. Citation2013). For phylogenetic analysis, we aligned the complete chloroplast genome sequences of nine species in the order Malpighiales including I. chinensis and Erythroxylum novogranatense, with Populus euphratica as an outgroup. The sequences were aligned using MAFFT (Katoh and Standle Citation2013), and a maximum likelihood tree was constructed with RAxML implemented in Geneious version.10.1 (Biomatters Ltd., Auckland, New Zealand; Kearse et al. Citation2012).
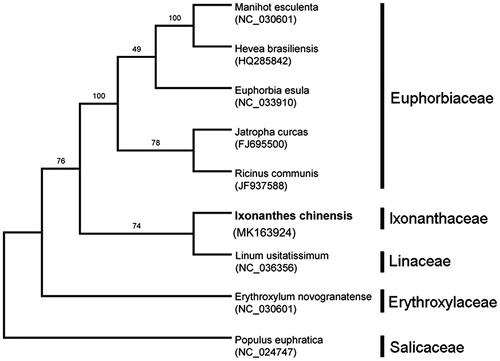
The complete chloroplast genome of I. chinensis is 155,296 bp in length. It consists of four typical regions: a large single copy (LSC) region of 83,829 bp and a small single copy (SSC) region of 18,917 bp, and separated by two inverted repeats (IRs) of 26,275 bp. The overall GC content is 36.91%. We predicted a total of 139 genes in this chloroplast genome, containing 87 protein-coding genes, 44 tRNA genes, and 8 rRNA genes. As shown in the phylogenetic tree (), I. chinensis is sister to Linum usitatissimum from the family Linaceae rather than Erythroxylum novogranatense from the family Erythroxylaceae. The chloroplast genome sequence would also be useful for investigating genetic diversity of I. chinensis in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Fu LK. 1992. China plant red data book: rare and endangered plants Vol.1. Beijing: Science Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Sun W. 1998. Ixonanthes chinensis. The IUCN red list of threatened species 1998: e.T32414A9704389. http://dx.doi.org/10.2305/IUCN.UK.1998.RLTS.T32414A9704389.en.
- Wu TL. 2000. Flora of Guangdong. Guangzhou: Guangdong Science and Technology Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3325.
- Xi ZX, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, Davis CC. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 109:17519–17524.