?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
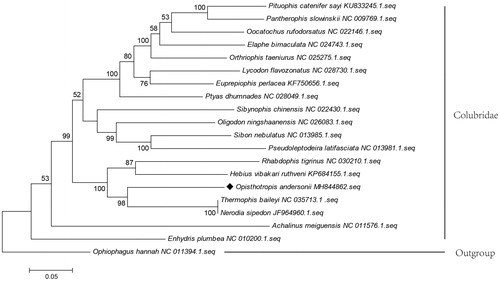
The complete mitochondrial genome (mitogenome) of Opisthotropis andersonii was first determined using illumina next-generation sequencing. The total length of genome was 18,297bp with a slightly AT bias (59.57%), and contained 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, 2 control regions, and an origin of light-strand replication. All genes are distributed on the H-strand, except ND6 gene. A phylogenetic tree was constructed based on the 13 PCG sequences of O. andersonii and other related snakes to analyze their phylogenic relationship, exhibiting the closest relationship with Thermophis baileyi and Nerodia sipedon.
Opisthotropis andersonii was listed as a near threatened species in the Red List (IUCN 2018) and was considered non-venomous, belonging to the genus Opisthotropis of family Colubridae, which was distributed in China and Vietnam (Zhao et al. Citation1998; David et al. Citation2011). There are 21 species in the genus Opisthotropis presently (Wang et al. Citation2017). But, the complete mitochondrial genome of these species is not published and few molecular data are available. In this paper, we determined and described the complete mitogenome of O. andersonii in order to obtain basic genetic information about this species, enrich the Opisthotropis species genome resource, and promote the further research concerning the related species.
Opisthotropis andersonii was collected from the Baiyun mount, Guangzhou, China. It was stored in the Specimen Museum of Guangdong Institute of Applied Biological Resources, GDAS. Whole genomic DNA was extracted from the specimen’s liver using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Genomic DNA was prepared in 150 bp paired-end libraries, tagged and subjected to the high-throughput Illumina Hiseq Xten platform, and yielded 29,750,352 raw reads. DNA sequences were assembled using MITObim version1.9 (Hahn et al. Citation2013). Comparing with the Nerodia sipedon (GenBank: JF964960.1), annotations were generated in MITOchondrial genome annotation Server (MITOS) (Bernt et al. Citation2013).
The total length of O. andersonii was 18,297 bp (GenBank: MH844862). The base composition of the complete mitogenome was 34.20% A, 13.14% G, 25.37% T, and 27.23% C, with a slightly A + T bias of 59.57%. The complete mitochondrial genome included 13 protein-coding genes(PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, 2 control regions (CRs), and an origin of light-strand replication (OL), which is similar to the typical mtDNA of reported snakes (Xu et al. Citation2015; Sun et al. Citation2017). All protein-coding genes had 11,888 bp length for coding 3730 amino acid. Twelve protein-coding genes were encoded on heavy strand (H-strand) and only ND6 gene on the light strand. Most protein-coding genes initiated with ATT or ATG except for ND4L and ND6, which began with GTG or ATA. Eight protein-coding genes terminated with the complete stop codon TAA (ND1, ND2, ATP8, ATP6, ND4L, ND4, ND5 and Cytb), ND6 regarded AGG as a stop codon, and the rest showed an incomplete stop codon T––. The size of the 22 tRNA genes ranged from 57 (tRNASer-AGY) to 73 bp (tRNALeu-UUR). The OL was 35 bp in length, located between tRNAAsn and tRNACys genes. The 12s rRNA and 16s rRNA were 924 and 1461 bp, respectively, separated by the tRNAVal.
To validate the phylogenetic relationship of O. andersonii within the Colubridae, the Maximum Likelihood tree was constructed by MEGA 7.0 (Kumar S et al. Citation2016) using 13 PCGs from O. andersonii, the other related 18 species were from the family Colubridae, and one outgroup from Elapidae (). The ML tree showed that O. andersonii clustered with the Thermophis baileyi and Nerodia sipedon, highly supported by a bootstrap value of 99%, and then clustered with Hebius vibakari ruthveni and Rhabdophis tigrinus, which was in accordance with recent molecular works (Wang et al. Citation2017). The mitogenome obtained in this study enriches the genomic resources available for further evolutionary studies, and can provide genomic resources for Cloubridae studies.
Disclosure statement
The authors report no conflicts of interest and are solely responsible for the content and the writing of the paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- David P, Pham TC, Nguyen QT, Ziegler T. 2011. A new species of the genus Opisthotropis Günther, 1872 (Squamata: Natricidae) from the highlands of Kon Tum Province. Vietnam Zootaxa. 2758:43–56.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Sun HJ, Li E, Sun L, Yan P, Xue H, Zhang F, Wu XB. 2017. The complete mitochondrial genome of the greater green snake Cyclophiopsmajor (Reptilia, Serpentes, Colubridae). Mitochondrial DNA Part B. 2:309–310.
- Wang YY, Guo Q, Liu ZY, Lyu ZT, Wang J, Luo L, Sun YJ, Zhang YW. 2017. Revisions of two poorly known species of Opisthotropis Günther, 1872 (Squamata: Colubridae: Natricinae) with description of a new species from China. Zootaxa. 4247:391–412.
- Xu CZ, Zhao S, Han XM. 2015. Sequence and organization of the complete mitochondrial genome of Hebius vibaka riruthveni from China. Mitochondrial DNA. 27:1–2.
- Zhao EM, Huang MH, Zong Y, Zheng J, Huang ZJ, Yang D, Li DJ. 1998. Fauna sinica. Reptilia. Vol. 3. Squamata serpentes. Beijing: Science Press. [in Chinese].