Abstract
Here, the complete mitogenome of three generations of ‘Rongfu’ were sequenced. The three genome sequences were exactly the same, whose circular genomes were all 37,638 bp in length, consisting of 35 protein-coding genes, 25 transfer RNA genes, 3 ribosomal RNA genes, and 3 open reading frames. The genome composition with A + T bias (64.66%) and gene arrangement was similar to those Saccharina species. The phylogenetic analysis showed that the three generations of ‘Rongfu’, ‘Rongfu’ in NCBI, and ‘Fujian’ in NCBI were clustered together, which will reveal the genetic and evolutionary characteristics at genomic level.
Saccharina (Phaeophyceae, Laminariales) is a large seaweed with important economic and ecological value (Jensen Citation1993; Kain Citation1979). ‘Rongfu’ (‘Fujian’♀ × ’Yuanza No.10’♂) is one of the most important commercially cultivar in China (Zhang, Li, et al. Citation2011; Zhang, Liu, et al. Citation2011). Here, the complete mitogenome of the three continuous generations of ‘Rongfu’ were obtained and compared with themselves and ‘Rongfu’ (JF937591) in NCBI. The phylogenetic tree was conducted to study the genetic stability of mitogenome in continuous generations and the genetic and evolutionary characteristics at genomic level.
Three ‘Rongfu’ samples (specimen number: 200905064, 2010081231, 201104101) were collected from Rongcheng, Shandong Province, China (37°15′39′N, 122°33′56′E), and stored at −80 °C in the Culture Collection of Seaweed at the Ocean University of China. The experimental methods and data analysis were followed by the previous reports (Li et al. Citation2018; Zhang, Li, et al. Citation2011).
The three genome sequences were found to be exactly the same through sequence alignment. Their mitochondrial genomes were all circular molecules with a length of 37,638 bp. The overall base composition of the mitogenomes were all found to be 28.40% A (10,689), 14.74% C (5549), 20.60% G (7754), and 36.26% T (13,646) with a high A + T content (64.66%). The AT skew (−0.1251) and GC skew (0.1658) indicated that H-strand had a slight base bias toward G and T, which was similar to other Saccharina species (Yotsukura et al. Citation2010; Zhang et al. Citation2013). The three mitogenomes encoded 66 genes which consisted of 35 protein-coding genes, 3 rRNA genes, 3 ORFs, and 25 tRNA genes. All protein-coding genes treated the typical initiation ATG as the start codon. About 68.42% protein-coding genes had TAA as termination codon and the remainings were TAG (21.05%), TGA (10.53%).
Compared with the complete mitogenomes of ‘Rongfu’ in NCBI, there were six nucleotide substitutions in the three generations in this research. Two of the substitutions were located in rRNA, and the remainings were respectively located in ORF377, cox2, nad5, and tatC. In total, there were no difference in the component and arrangement of mitogenome between the previous reported ‘Rongfu’ in NCBI (Zhang, Li, et al. Citation2011) and the three generations of ‘Rongfu’ in this research, showing high genetic stability of mitogenome of ‘Rongfu’.
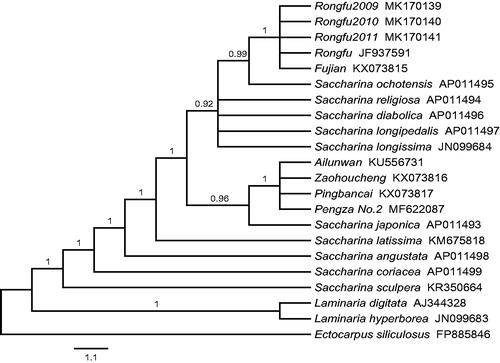
The phylogenetic tree was constructed based on 35 protein-coding genes from 21 available Laminariaceae species, with Ectocarpus siliculosus used as the outgroup, by Bayesian method. As shown in the figure, 22 species were separated into two clades: Saccharina and Laminaria, and all of the cultivars in China were in Saccharina clade (). The cultivars ‘Ailunwan’, ‘Zaohoucheng’, ‘Pingbancai’, and ‘Pengza No.2’ were clustered with S. japonica which supported their parental origin. Due to the lack of non-coding region of 19 nucleotides, the three generations of ‘Rongfu’, ‘Rongfu’ in NCBI, and ‘Fujian’ (the female parent of ‘Rongfu’) in NCBI were clustered together and formed a separate cluster from other cultivars.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jensen A. 1993. Present and future needs for algae and algal products. Hydrobiologia. 260–261(1):15–23.
- Kain JM. 1979. A view on the genus Laminaria. Oceanogr Mar Biol Annu Rev. 17:101–161.
- Li Y, Liu N, Yin HX, Liu C, Zhang L, Jin YM, Wang HY, Chi S, Liu T. 2018. Complete sequences of the mitochondrial DNA of the Petalonia binghamiae. Mitochondrial DNA B. 3(1):95–96.
- Yotsukura N, Shimizu T, Katayama T, Druehl LD. 2010. Mitochondrial DNA sequence variation of four Saccharina species (Laminariales, Phaeophyceae) growing in Japan. J Appl Phycol. 22(3):243–251.
- Zhang J, Li N, Zhang Z, Liu T. 2011. Structure analysis of the complete mitochondrial genome in cultivation variety ‘Rongfu’. J Ocean Univ China. 10(4):351–356.
- Zhang J, Liu Y, Yu D, Song H, Cui J, Liu T. 2011. Study on high-temperature-resistant and high-yield Laminaria variety “Rongfu”. J Appl Phycol. 23(2):165–171.
- Zhang J, Wang X, Liu C, Jin Y, Tao L. 2013. The complete mitochondrial genomes of two brown algae (Laminariales,Phaeophyceae) and phylogenetic analysis within Laminaria. J Appl Phycol. 25(4):1247–1253.