Abstract
The complete mitochondrial genome of a herbivorous amphipod, Ampithoe lacertosa, has been determined for the first time in the present study. The total mitogenome length of A. lacertosa was 14,607 bp with 13 protein-coding genes, two ribosomal RNA genes, and 22 transfer RNA genes. Phylogenetic analysis showed that A. lacertosa belonged to the families Caprellidae, Hyalellidae, Hyalidae, and Talitridae in the same clade, and to the suborder Senticaudata within Amphipoda. This is the first record of the complete mitochondrial genome sequence of the family Ampithoidae.
Ampithoidae Boeck, 1871, a group of herbivorous amphipods, is an important invertebrate member that dominates invertebrate assemblages associated with macroalgae and seagrasses in shallow coastal waters worldwide (Myers and Lowry Citation2003; Shin et al. Citation2015; Sotka et al. Citation2017). The genus Ampithoe Leach, 1814 is the largest member of the family Ampithoidae, and so far 80 species have been reported (Horton et al. Citation2018). In this study, we provide the complete mitogenome sequence of an ampithoid species, Ampithoe lacertosa Spence Bate (Citation1858).
The specimen was collected from the tidal zone in the southern coast of East Sea, Gyeong Ju, Korea (35°48′25.7′′N, 129°30′32.6′′E), on July 11th 2018. The voucher specimen was deposited at the National Marine Biodiversity Institute of Korea. Subsequent to mtDNA enrichment procedures (mitochondrial isolation, extraction and amplification of mtDNA), mtDNA was extracted and sequenced using Illumina HiSeq2500 platform. Short-read DNA sequences were assembled and analyzed using Geneious 9.0.5 (Biomatters). The 13 protein-coding genes were annotated by identifying their open reading frames, and by comparing them with other reported mitogenomes of amphipods using DOGMA (Wyman et al. Citation2004) and ORF finder in the NCBI web server. All transfer RNAs were identified with tRNAscan-SE 1.21 (Schattner et al. Citation2005) and ARWEN 1.2 (Laslett and Canbäck Citation2007), which inferred cloverleaf secondary structures. The ribosomal RNA genes were identified by sequence comparison with other reported mitogenomes of amphipods.
The complete mitogenome of A. lacertosa was 14,607 bp in length (GenBank accession no. MK215645). It encoded 37 genes, 13 protein-coding genes, two rRNA genes, and 22 tRNA genes, with a non-coding region of 616 bp. The overall AT content of the mitogenome was 77% (39.3% A, 9.8% C, 13.1% G, and 37.7% T). There were four types of PCGs start codons, ATT (cox1–2, nad2, nad4–5, and atp8), ATA (nad6), ATG (atp6, cytb, cox3, nad3, and nad4L), and TTG (nad1). TAA (cox3, atp6, atp8, nad3, nad4L, and nad6) and TAG (nad1) were stop codons. Furthermore, the nucleotide ‘A’ which was located at the end of some PCGs (cox1–2, atp8, cytb, nad2, and nad4–5), was used to avoid overlapping between the genes.
To confirm the phylogenetic position of A. lacertosa, another eleven species belonging to six families in the suborder Senticaudata were used, with a lysianassid species of the suborder Amphilochidea as an outgroup (all the species were registered in the NCBI). A phylogenetic tree was reconstructed based on the concatenated data set of 13 PCGs, using the neighbour-joining method with MEGA7 (Kumar et al. Citation2016). As a result, A. lacertosa was grouped with another four families, Caprellidae, Hyalellidae, Hyalidae, and Talitridae, in one clade with high bootstrap value, and it belonged to the suborder Senticaudata within Amphipoda ().
Figure 1. A phylogenetic tree built using a concatenated data set of 13 protein-coding genes by the neighbor-joining (NJ) method, with the Kimura 2-parameter model. The tree was reconstructed based on 13 mitogenome sequences, including that of Ampithoe lacertosa which was examined in the present study. Bootstrap values were calculated from 1,000 replicates. The herbivorous amphipod, Ampithoe lacertosa is highlighted in black.
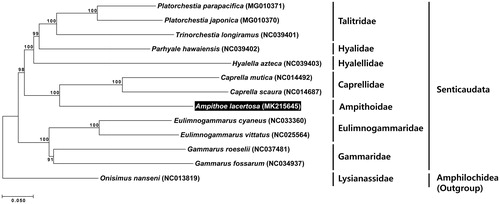
This is the first record complete mitogenome sequence of the family Ampithoidae. The results of the present study provide useful information for further phylogenetic and evolutionary studies on amphipod members in the future.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bate CS.. 1858. On some new genera and species of Crustacea Amphipoda. Ann. Mag. Nat. Hist. ser 3.. 1:361–362.
- Horton T, Lowry J, De Broyer C, Bellan-Santini D, Coleman CO, Corbari L, Costello MJ, Daneliya M, Dauvin J-C, Fišer C, et al. 2018. Ampithoe Leach, 1814. [Ostend (BE)]: World Amphipoda Database. [accessed 2018 Nov 7]. http://www.marinespecies.org/aphia.php?p=taxdetails&id=101459.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol.33(7):1870–1874.
- Laslett D, Canbäck B. 2007. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Myers AA, Lowry JK. 2003. A phylogeny and a new classification of the Corophiidea Leach, 1814 (Amphipoda). J Crust Biol. 23:443–485.
- Shin MH, Coleman CO, Hong JS, Kim W. 2015. A new species of Peramphithoe (Amphipoda: Ampithoidae) from South Korea, with morphological diagnoses of the world congeneric species. J Crust Biol. 35:255–270.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. J Nucleic Acids. 33:686–689.
- Sotka EE, Bell TL, Hughes E, Lowry JK, Poore AGB. 2017. A molecular phylogeny of marine amphipods in the herbivorous family Ampithoidae. Zool Scr. 46:85–95.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
