Abstract
In the present study, the complete mitogenome of a dorid nudibranch Phyllidiella pustulosa was sequenced and analyzed. The complete mitochondrial genome of P. pustulosa was 14,717 bp in length, containing 13 protein coding genes, two ribosomal RNA genes, and 22 tRNA genes. Phylogenetic analysis according to protein coding genes revealed that P. pustulosa has a close relationship with Phyllidia ocellata. This is the first complete mitochondrial genome record for the genus Phyllidiella.
Phyllidiella pustulosa is a common phyllid nudibranch distributed in the Indo-West Pacific region. Characteristics for morphological identification include elongated, ovate body shape and pink clustered tubercles on a black dorsum (Brunckhorst Citation1993). However, a phylogenetic analysis based on partial cytochrome C oxidase subunit I (cox1) gene has indicated that P. pustulosa is potentially a species complex (Stoffel et al. Citation2016). The mitochondrial genome with multiple markers is a useful tool for species identification. Therefore, the complete mitogenome of P. pustulosa might be helpful to discriminate its complex.
The P. pustulosa sample was collected in May 2013 from Dodu-dong, Jeju island, Korea (33°31′19.1″ N – 126°27′04.3″ E) through scuba diving. The sample was preserved in 97% ethanol and deposited in the Department of Biotechnology, Sangmyung University, Korea (SMU00012) for further analysis. The DNA of P. pustulosa was extracted from the foot of the specimen using E.Z.N.A.® Mollusc DNA Kit (Omega Bio-tek, Norcross, GA, USA). Paired end reads of mitogenome were generated by Illumina Miseq sequencing system (San Diego, CA, USA). The reads were targeted for reconstructing the complete mitogenome by MITObim (Hahn et al. Citation2013). The circular sequence was annotated using MITOS web server (Bernt et al. Citation2013) and Geneious 9.1 software (Kearse et al. Citation2012).
The size of the complete mitogenome of P. pustulosa (GenBank accession number: MK279705) was 14,717 bp. Similar to the other nudibranchs, it contained 13 protein-coding genes (PCGs), 22 transfer RNA genes, and 2 ribosomal RNA genes. Nucleotide content of the mitogenome consisted of 30.3% A, 15.1% C, 17.5% G, and 37.1% T. The longest protein coding gene was nd5 (1683 bp) while the shortest protein coding gene was atp8 (156 bp). The sizes of 12S rRNA and 16S rRNA genes were 758 bp and 1244 bp, respectively. The tRNA gene lengths ranged from 56 bp (tRNA-Ser 2) to 71 bp (tRNA-Arg and tRNA-Cys).
Among PCGs, ten genes started with ATG codon, two genes (nd3 and nd4l) started with ATA codon, and one gene (nd6) started with ATT codon. It should be noted that there were 11 genes terminated with TAA and only two genes (cox3 and nd5) terminated with TAG. In total, 13 genes were encoded on L-strand, including four PCGs (atp6, atp8, cox3, and nd3), one rRNA gene (12S rRNA) and eight tRNA genes (tRNA-Gln, tRNA-Leu 2, tRNA-Asn, tRNA-Arg, tRNA-Glu, tRNA-Met, tRNA-Ser 2, and tRNA-Thr).
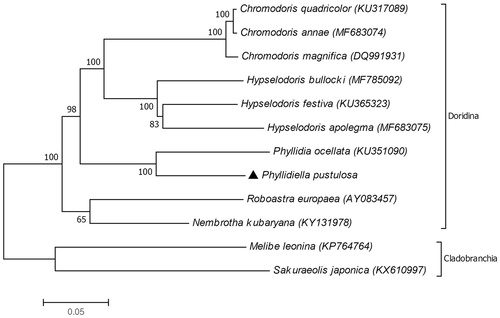
The phylogenetic position of P. pustulosa was constructed based on amino acid sequences of 12 PCGs (atp8 gene excluded). The result showed that P. pustulosa formed a clade with Phyllidia ocellata in the suborder Doridina (). The previous molecular phylogeny study based on 16S rRNA and cox1 gene suggested the similar relationship between the Phyllidiella and Phyllidia genera (Stoffel et al. Citation2016). In addition, phylogenetic tree according to cox1 from the previous study demonstrated that there are different clades inside P. pustulosa (Stoffel et al. Citation2016). Our study provides the mitogenome of P. pustulosa from Korean waters. To confirm the P. pustulosa species complex, more mitogenomes from different geographic localities should be added.
Figure 1. Molecular phylogeny of Phyllidiella pustulosa and other related species in the suborder Doridina. The complete mitochondrial genomes of nudibranchs were obtained from the GenBank. Melibe leonia and Sakuraeolis japonica of the suborder Cladobranchia were used as outgroup. Phylogenetic tree based on concatenated amino acid sequences of 12 protein coding genes (atp8 excluded) was constructed by neighbor-joining method along with 1000 bootstrap replicates in MEGAX software (Kumar et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brunckhorst DJ. 1993. The systematics and phylogeny of phyllidiid nudibranchs (Doridoidea). Rec Aust Mus, Suppl. 16:1–107.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Stoffel BEMW, van der Meij SET, Hoeksema BW, van Alphen J, van Alen T, Meyers-Muñoz MA, de Voogd NJ, Tuti Y, van der Velde G. 2016. Phylogenetic relationships within the Phyllidiidae (Opisthobranchia, Nudibranchia). Zookeys. 605:1–35.
