Abstract
Pseudostellaria palibiniana (Takeda) Ohwi which belongs to Caryophyllaceae is distributed in Korea and Japan. In this study, we present the complete chloroplast genome of P. palibiniana which is 149,668 bp long and has four subregions: 81,313 bp of large single copy (LSC) and 16,958 bp of small single copy (SSC) regions are separated by 25,709 bp of inverted repeat (IR) regions including 126 genes (81 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.5% and those in the LSC, SSC, and IR regions are 34.3%, 29.4%, and 42.4%, respectively.
Pseudostellaria palibiniana (Takeda) Ohwi is a perennial and herbaceous plant which is only distributed in the Korean peninsula and Honshu in Japan (Ōi et al. Citation1965; Park Citation2007). It also contains secondary metabolites which have pharmacological effect (Yang and Kim Citation1984) like Pseudostellaria heterophylla (Han et al. Citation2008; Chen et al. Citation2018; Deng et al. Citation2018). It lives mainly in foothills and forests. This species adopts a mixed-mating system which has both open-pollinated chasmogamous and permanently closed self-pollinating cleistogamous flowers (Culley and Klooster Citation2007). P. palibiniana was classified in series Mamillatae by its seed coat and in subseries Verticillatae by leaf arrangement (pseudo whorl; Ohwi Citation1937). Despite obvious morphological differences among four species in subseries Verticillatae, P. palibiniana is unresolved with the other species, named as P. palibiniana species complex (Kim et al., in preparation).
We sequence complete chloroplast genome of P. palibiniana to understand the phylogenic relationship of Pseudostellaria species with the already published two chloroplast genomes of Pseudostellaria genus (Kim and Park Citation2009; Kim et al. Citation2018). Pseudostellaria palibiniana was collected in Mt. Gwangdeok, Gangwon Province, Korea (N38°7'18.68", E 127°26'14.23"). The voucher and its DNA are deposited in InfoBoss Cyber Herbarium (IN; Y. Kim, IB-00588). Total DNA was extracted from fresh leaves of P. palibiniana by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq2000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and confirmed all assembled bases using BWA 0.7.17 and SAMtools 1.9 (Li et al. Citation2009; Li Citation2013). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on Pseudostellaria longipedicellata (Kim et al. Citation2018).
The chloroplast genome of P. palibiniana (Genbank accession is MK120981) is 149,626 bp long and has four subregions: 81,313 bp of large single copy (LSC) and 16,958 bp of small single copy (SSC) regions are separated by 25,709 bp of the inverted repeat (IR). It contains 126 genes (81 protein-coding genes, eight rRNAs, and 37 tRNAs); 18 genes (seven protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions. The overall GC content of P. palibiniana is 36.5% and those in the LSC, SSC, and IR regions are 34.3%, 29.4%, and 42.4%, respectively.
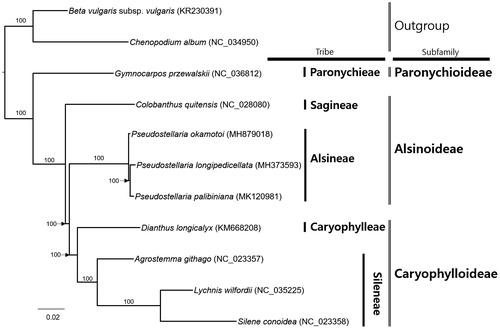
Nine complete chloroplast genomes of Caryophyllaceae including three Pseudostellaria species and two species from Amaranthaceae were used for constructing maximum likelihood phylogenetic tree. Alignment was conducted by MAFFT 7.388 (Katoh and Standley Citation2013) and a phylogenetic tree was constructed by IQ-TREE 1.6.6 (Nguyen et al. Citation2015). The phylogenetic tree shows that three Pseudostellaria species belonging to P. palibiniana species complex (Kim et al., in preparation) is resolved clearly with monophyletic manner (). The tree also presents that Alsinoideae, containing Alsineae and Sagineae, is paraphyletic, which is different from the previous molecular phylogenic study (Greenberg and Donoghue Citation2011; ). In consequence, Caryophylloideae and Alsinoideae are monophyletic (Greenberg and Donoghue Citation2011); while the tree shows that Caryophylloideae is monophyletic but Alsinoideae is paraphyletic (). Our results indicate that Pseudostellaria chloroplast genomes can provide additional information on phylogenetic relationships in Caryophyllaceae.
Figure 1. Maximum likelihood phylogenetic tree of Caryophyllaceae based on 11 complete chloroplast genomes: Pseudostellaria palibiniana (MK120981; in this study), Pseudostellaria okamotoi (MH879018); Pseudostellaria longipedicellata (MH373593), Beta vulgaris subsp. vulgaris (KR230391), Chenopodium album (NC_034950), Gymnocarpos przewalskii (NC_036812), Colobanthus quitensis (NC_028080), Dianthus longicalyx (KM668208), Agrostemma githago (NC_023357), Lychnis wilfordii (NC_035225), and Silene conoidea (NC_023358). The numbers above branches indicate bootstrap support values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Culley TM, Klooster MR. 2007. The cleistogamous breeding system: a review of its frequency, evolution, and ecology in angiosperms. The Bot Rev. 73:1.
- Chen J, Pang W, Kan Y, Zhao L, He Z, Shi W, Yan B, Chen H, Hu J. 2018. Structure of a pectic polysaccharide from Pseudostellaria heterophylla and stimulating insulin secretion of INS-1 cell and distributing in rats by oral. Int J Biol Macromol. 106:456–463.
- Deng Y, Bangxing H, Dejun H, Jing Z, Shaoping L. 2018. Qualitation and quantification of water soluble non-starch polysaccharides from Pseudostellaria heterophylla in China using saccharide mapping and multiple chromatographic methods. Carbohydrate Polymers. 199:619–627.
- Greenberg AK, Donoghue MJ. 2011. Molecular systematics and character evolution in Caryophyllaceae. Taxon. 60:1637–1652.
- Han C, Shen Y, Chen J, Lee FS-C, Wang X. 2008. HPLC fingerprinting and LC-TOF-MS analysis of the extract of Pseudostellaria heterophylla (Miq.) Pax root. J. Chromatogr. B Analyt Technol Biomed Life Sci. 862:125–131.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kim Y, Park J. 2019. Complete chloroplast genome sequence of the Pseudostellaria okamotoi Ohwi (Caryophyllaceae). Mitochondrial DNA Part B. 4:174–175.
- Kim Y, Heo K-I, Lee S, Park J. 2018. Complete chloroplast genome sequence of the Pseudostellaria longipedicellata S. Lee, K. Heo & SC Kim (Caryophyllaceae). Mitochondrial DNA Part B. 3:1296–1297.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Ohwi J. 1937. The Revision of the genus Pseudostellaria. Jap J Bot. 9:95–105.
- Ōi J, Meyer FG, Walker EH. 1965. Flora of Japan. Washington, USA: Smithsonian Institution.
- Park C. 2007. The genera of vascular plants of Korea. Seoul: Academic Publishing 1482p ISBN. 745696222.
- Yang K-S, Kim T-H. 1984. Pharmacological Studies on Pseudostellaria palibiniana. Kor J Pharmaco. 15:6–14.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Gen Res. 18:821–829.
