Abstract
Cephalotaxus hainanensis Li is a vulnerably endangered gymnosperm species with medicinal value. Here we performed the chloroplast (cp) genome of C. hainanensis by next generation sequencing to provide the potential information for conservation and genetic studies. The cp genome was 136,115 bp length and the overall GC content was 36.06%. It encodes 115 unigue genes, including 83 protein coding, 28 tRNA, and 4 rRNA genes. A neighbour-joining phylogenetic analysis suggested a close relationship between C. hainanensis and C. sinensis.
Cephalotaxus hainanensis Li, a tertiary relict tree circumscribed in the genus of Cephalotaxus (Cephalotaxaceae), is endemic to Hainan Island of China (Fu et al. Citation1999). The bark of this tree produces special alkaloids which are utilized as a medicine for acute myeloid leukemia and malignant lymphoma (Mei et al. Citation2006; Abdelkafi and Nay Citation2012). Due to its growth slowly and overharvesting for medical purpose, C. hainanensis has been classified as an endangered gymnosperm species in China and listed in the IUCN Red List from 2013 (Qiao et al. Citation2014). There is an essential need for alternatives and incentives for conservation of this threatened species. Previous studies about C. hainanensis were focused on chemicals constituents and pharmacology. However, there have been no reports about chloroplast genome information of C. hainanensis until now. In this study, we determined the complete chloroplast genome of C.hainanensis using high throughput sequencing technology, which will provide informatics data for the phylogeny of Cephalotaxus genus and the conservation of C. hainanensis.
Fresh leaves were collected from an individual tree of C. hainanensis in Bawang mountain (Changjiang, Hainan Province, China). The total genomic DNA was extracted using a Plant Genomic DNA Kit (Tiangen, Beijing, China) and then stored in the herbarium of our institute. Subsequently, the genomic DNA was used to generate 440bp (insert size) library according to the standard protocol of Illumina Hiseq X Ten (Illumina, CA, USA). Approximately 4.06 Gb raw data were obtained with pair-end 150 bp read length and these raw data were filtered by using NGS QC Toolkit_v2.3.3 for high-quantity clean reads. The cp genome was assembled by using SOAPdenovo v2.04 and this draft cp genome was subsequently corrected based on GapCloser v1.12. The annotation was performed using DOGMA and then submitted to GenBank (accession no. MK125083).
The complete C.hainanensis cp genome is 136,115 bp and its base composition was asymmetric (32.63% A, 32.31% T, 17.58% C, 17.48% G) with an overall GC content of 36.06%. The cp genome of C. hainanensis contained 115 genes, including 83 protein-coding genes, 28 transfer RNA genes, and 4 ribosomal RNA genes. Nine genes contained a single intron, whereas two genes have two introns. Nevertheless, the large single copy (LSC) and small single copy (SSC) regions could not be classified for lack of the typical inverted repeats (IRs) regions in cp genome of C. hainanensis. This result is consistent with C. wilsoniana and C. oliveri cp genome from the congeneric Cephalotaxus and Pinaceae plant (Wu et al. Citation2011; Yi et al. Citation2013), which provides new evidence for the loss of IR regions in cp genome evolution.
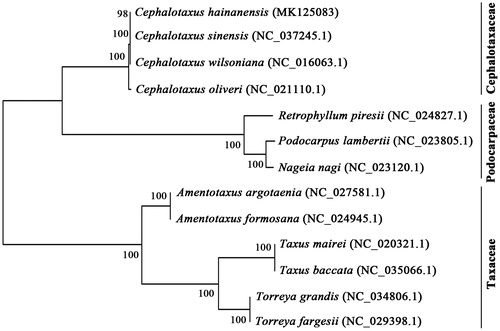
To ascertain phylogenetic position of C. hainanensis within Taxopsida, a neighbour-joining (NJ) phylogenetic tree was constructed with MEGA X (Kumar et al. Citation2018) using whole cp genome sequences of 13 representatives in Taxopsida (). The result indicated that the close relationship between C. hainanensis and C. sinensis, while both of which are clustered together with C. wilsoniana and C. oliveri in genus of Cephalotaxus, the sole representative of the family Cephalotaxacea.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abdelkafi H, Nay B. 2012. Natural products from Cephalotaxus sp.: chemical diversity and synthetic aspects. Nat Prod Rep. 29:845–869.
- Fu L, Li N, Mill RR. 1999. Cephalotaxaceae. Flora China. 4:85–88.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. Mega x: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Mei WL, Wu J, Dai HF. 2006. Advances in studies on chemical constituents in plants of Cephalotaxus Sieb. et Zucc. and their pharmacological activities. Chin Tradit Herb Drug. 93:219–220.
- Qiao F, Cong H, Jiang X, Wang R, Yin J, Qian D, Wang Z, Nick P. 2014. De novo characterization of a Cephalotaxus hainanensis transcriptome and genes related to paclitaxel biosynthesis. PLoS One. 9:e106900.
- Wu CS, Wang YN, Hsu CY, Lin CP, Chaw SM. 2011. Loss of different inverted repeat copies from the chloroplast genomes of pinaceae and cupressophytes and influence of heterotachy on the evaluation of gymnosperm phylogeny. Genome Biol Evol. 36:1284–1295.
- Yi X, Gao L, Wang B, Su Y-J, Wang T. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biol Evol. 5:688–698.