Abstract
In this study, the complete mitochondrial genome of smoothshell shrimp Parapenaeopsis tenella was determined for the first time. The mitogenome of P. tenella is 15,893 bp in length and encodes 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region (CR). The overall base composition is 34.0%, 35.3%, 11.9%, and 18.9% for A, T, C, and G, respectively. All PCGs are initiated by ATN codons. Four of the 13 PCGs harbor the incomplete termination codon by T. Twenty-one tRNA genes have a typical clover-leaf structure of mitochondrial tRNA, except for tRNA-Ser (AGC), in which the dihydrouridine (DHU) arm stem is absent. Phylogenetic analysis among the available malacostracans species supports the current morphology-based hypothesis that P. tenella is grouped to the genus Parapenaeopsis.
The genus Parapenaeopsis is one of the natural group of small-sized shrimp inhibiting mainly in the shallow coastal areas of tropical and subtropical seas in the Indo-West Pacific region (Liu and Wang Citation1987). Many species of this genus, such as Parapenaeopsis hardwickii and P. tenella, are of great importance in marine fishery in China. However, at present, genetic information available for smoothshell shrimp P. tenella is quite limited, which constrains population genetics study for this species. In the present study, the complete mitochondrial genome of P. tenella was determined for the first time.
The specimen of P. tenella was collected from Yellow Sea of China (33.011932°N, 122.015328°E), and stored at −80 °C prior to DNA extraction. DNA was isolated with the phenol–chloroform method as described in Santos (Citation2006). All the specimen and DNA samples were deposited in the sample room of Division of Genetic Resources and Breeding, Yellow Sea Fisheries Research Institute. The mitogenome of P. tenella was amplified with 19 pairs of primers. Phylogenetic analysis was carried out using the maximum likelihood method with MEGA7.0 software (Kumar et al. Citation2016).
The mitogenome of P. tenella is 15,893 bp in length and is deposited in GenBank under Accession No. MK164420. The A–T content of the P. tenella mitogenome is 69.3%, containing 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and 1 control region (CR). The CR is 997 bp in length and is located between the 12S rRNA and tRNA-Ile genes. The two mitochondrial rRNA genes of P. tenella are both located on the light strand. Nine of the 13 PCGs are encoded on the heavy strand while the remaining four are located on the light strand, which is relatively universal across mitochondrial genomes of the malacostracan species sequenced to date (Mao et al. Citation2016; Meng et al. Citation2016). All the PCGs in the P. tenella mitochondrial genome are initiated with ATN. Nine of the PCG genes have a complete termination codon either TAA or TAG, while the other PCGs have incomplete terminate codons, T, which were believed to be converted to complete TAA codons by the addition of A residues during RNA processing (Anderson Citation1982). The tRNA genes range from 64 to 72 bp in size. Most of the P. tenella tRNAs have typical cloverleaf structures, except for tRNA-Ser (AGN), which is also found in many other arthropod mitochondrial genomes (Kim et al. Citation2018; Zhong et al. Citation2018).
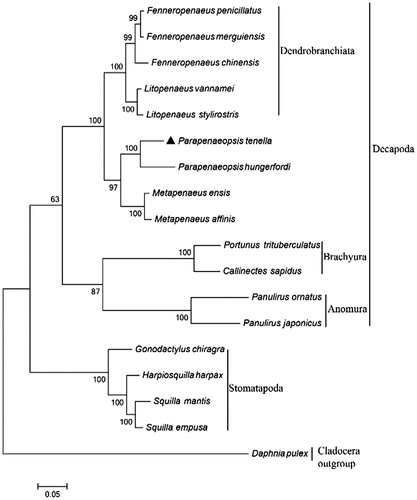
The phylogenetic position of P. tenella within Decapoda was investigated with the concatenated alignments of amino acid in 18 decapod species using the maximum likelihood method (). The results showed that P. tenella clustered with P. hardwickii and P. hungerfordi, which was consistent with the previous classification based on morphology (Sakai and Shinomiya, Citation2011). The P. tenella mitogenome resource obtained in the present study forms a valuable asset for improving the conservation of P. tenella via further investigations of population genetics.
Figure 1. Phylogenetic tree of Parapenaeopsis tenella and other crustacean species constructed using the maximum likelihood method with concatenated mitochondrial PCGs. Daphnia pulex in Cladocera was adopted as the outgroup member. Numbers below each node indicated the ML bootstrap support values. The GenBank accession numbers of complete mitochondrial genomes used in this phylogeny analysis are as follows: Fenneropenaeus chinensis (DQ656600.1), Fenneropenaeus merguiensis (KP637168.1), Fenneropenaeus penicillatus (KP637169.1), Litopenaeus stylirostris (EU517503.1), Litopenaeus vannamei (EF584003.1), Parapenaeopsis tenella (MK164420), Parapenaeopsis hungerfordi (NC_038069.1), Metapenaeus affinis (NC_039179.1), Metapenaeus ensis (NC_026834.1), Portunus trituberculatus (AB093006.1), Callinectes sapidus (AY363392.1), Panulirus japonicus (NC_004251.1), Panulirus ornatus (GQ223286.1), Gonodactylus chiragra (DQ191682.1), Harpiosquilla harpax (AY699271.1), Squilla empusa (DQ191684.1), Squilla mantis (NC_006081.1), and Daphnia pulex (KT003819.1).
Acknowledgements
We would like to thank Dr. Qiang Wu from the Division of Fishery Resources and Ecosystem, Yellow Sea Fisheries Research Institute, for collecting the sample and identifying its species.
Disclosure statement
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Additional information
Funding
References
- Anderson DT. 1982. Embryology: embryology, morphology and genetics. New York: Academic Press.
- Kim NK, Andriyono S, Kim AR, Lee CI, Kim HW. 2018. Characterization of complete mitochondrial genome of two-spot swimming crab Charybdis bimaculata (Miers, 1886). Mitochondrial DNA Part B. 3:902–903.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu RY, Wang YL. 1987. Studies on Chinese species of the genus Parapenaeopsis (Decapoda, Crustacea). Oceanol Limnol Sin. 18:523–539.
- Mao ZC, Liu P, Duan YF, Li J, Chen P, Meng XL. 2016. Sequencing of complete mitochondrial genome of sword prawn parapenaeopsis hardwickii (Miers) (Crustacea, Decapoda, Penaeidae). Mitochondrial DNA Part B. 1:259–260.
- Meng XL, Jia FL, Zhang XH, Liu P, Li J. 2016. Complete sequence and characterization of mitochondrial genome in the swimming crab Portunus sanguinolentus (Herbst, 1783) (Decapoda, Brachyura, Portunidae). Mitochondrial DNA Part A. 27:3052–3053.
- Sakai K, Shinomiya S. 2011. Preliminary report on eight new genera formerly attributed to Parapenaeopsis Alcock, 1901, sensu lato (Decapoda, Penaeidae). Crustaceana. 84:491–504.
- Santos SR. 2006. Patterns of genetic connectivity among anchialine habitats: a case study of the endemic Hawaiian shrimp Halocaridina rubra on the Island of Hawaii. Mol Ecol. 15:2699–2718.
- Zhong SP, Zhao Y, Wang XF, Song ZF, Zhang Q, Chen XL. 2018. The complete mitochondrial genome of the cryptic species (Form II) in kuruma shrimp Marsupenaeus japonicus (Decapoda: Penaeidae). Mitochondrial DNA Part B. 3:184–186.
