Abstract
Whole mitochondrial genome sequence of Apriona swainsoni has a circular genome of 15,412 bp. The contents of A, G, T, and C are 38.90%, 9.29%, 35.81%, and 15.99%, respectively. The proportion of coding sequences with a total length of 11, 141 bp is 72.29%, which encodes 3689 amino acids. Most protein-coding genes stopped with TAN, with COX1 and COX2 taking the incomplete codon T as the stop codon. The incomplete stop codon (TA and T) are ubiquitous in insect mitochondrial genome. The length of D-loop is 735 bp, ranging from 14,677 to 15,412 bp.
Apriona swainsoni is one of the important pests in China’s forest quarantine. It is also a serious pest on Sophora japonica (Liu Citation2002; Liu et al. Citation2006). One generation of Apriona swainsoni was 2 years, and it overwintered in the xylems of host trees in different larvae stages (Tang et al. Citation2001). This insect only did harm to Sophora japonica Linn. The toxic to stomach or contact insecticide used for the treatment of the tree trunk world was better for the control of the insect from middle to late June (Duan Citation2001). The optimum time foe control is when adults move about on trees (Liu and Zhang Citation2002). The insects were collected on Hibiscus syriacus from Meiling Aera, Nanchang, China(locating at E115°43′44″, N28°43′18″). Specimens were then stored in −80 °C refrigerators of the laboratory in Jiangxi Academy of Forestry.
Here, we sequenced and characterized the complete mt genome of Apriona swainsoni. The mtDNA sequence of Apriona swainsoni with the annotated genes was deposited in GenBank under the accession number of NC_033872. The overlapping sequences were manually checked and assembled into a complete mitogenome via the alignment of neighboring fragments using the Clustal X program (Thompson et al. Citation1997). Whole mitochondrial genome sequence of Apriona swainsoni has a circular genome of 15,412 bp, containing 13 protein-coding genes, 2 rRNA genes, 1 control region, and 22tRNA genes. The contents of A, G, T, and C are 38.90%, 9.29%, 35.81%, and 15.99%, respectively. AT and GC contents of mt genome are 74.71% and 25.29%, A + T is much higher than G + C. respectively. The proportion of coding sequences with a total length of 11,141 bp is 72.29%, which encodes 3689 amino acids. All protein-coding genes started with ATN. ND2, ATP6, COX3, ND5, ND4L, ND6, and Cytb ended by TAA as the stop codon. Remarkably, ATP8, ND3, and ND1 ended by TAG as the stop codon. Only COX1 and COX2 take the incomplete codon T as the stop codon. Incomplete stop codon (TA and T) are ubiquitous in insect mitochondrial genome. The lengths of 12S ribosomal RNA and 16S ribosomal RNA are 807 bp and 1287 bp, respectively. The length of D-loop is 735 bp, ranging from 14,677 to 15,412 bp.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of Apriona swainsoni and the other 19 species using as Diaphorina citri an outgroup. Number above each node indicates the ML bootstrap support values.
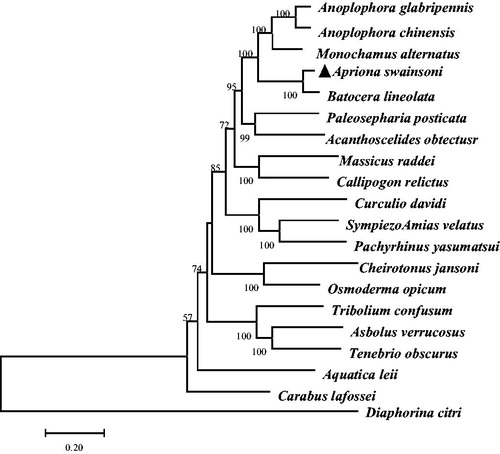
Phylogenetic analysis included mt genome of Apriona swainsoni and the other 19 species that are from the order Curculionidae, Chrysomelidae, Cerambycidae, Tenebrionidae, Carabidae, Lampyridae, Scarabaeidae, Lucanidae, Elateridae, which belong to Coleoptera (see ). Using Diaphorina citri (Kuwayama) of Psylloidea Family as an outgroup. Maximum likelihood (ML) methods were used for phylogenetic analysis. The GTR + G + I model was selected by the Akaike information criterion in MEGA 6.06 (Tamura et al. Citation2013). The obtained phylogenetic trees were visualized with Treeview 1.6.6 (Page Citation1996).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Duan Y. 2001. Study on Apriona swainsoni hope of devastating pest in Sophora japonica Linn[J]. J Anhui Agricuiturai Sciences. 29:375–377.
- Liu Y. 2002. Study on the spatial distribution patterns of the larvae of Apriona swainsoni Hope and its applications [J]. J Anhui Agricultural University. 29:233–236.
- Liu H, Luo Y, Wen J, Zhang Z, Feng J, Tao W. 2006. Pest risk assessment of Dendroctonus valens, Hyphantria cunea and Apriona swainsoni in Beijing[J]. Front Forest China. 1:328–335.
- Liu Y, Zhang L. 2002. Experiment on the control of Apriona swainsoni (Hope) [J]. Forest Pest and Dis. 2:32–34.
- Page RD. 1996. TreeView: an application to display phylogenetic trees on personal computers [J]. Comput Appl Biosci. 12:357–358.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0[J]. Mol Biol Evol. 30:2725–2729.
- Tang Y, Liu Y, Li X, et al. 2001. Investigation on biological characteristics of Apriona swainsoni Hope[J]. J Anhui Agricultural University. 28:238–241.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nuc Acids Res. 25:4876–4882.
