Abstract
The complete mt genome sequence of Pelteobagrus vachelli was obtained by PCR, containing 37 genes with 13 protein-coding genes, 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a non-coding control region.
Morphological classification of Bagridae fishes is relatively difficult due to various classifications and similar shape. In this study, the complete mitochondrial genome of P. vachelli has been sequenced.
Pelteobagrus vachelli was captured in Huai River (32°36′47.1″N, 116°26′19.2″E). The specimen was stored in Fihseries Institute, Anhui Academy of Agriculture Sciences. We amplified mitochondrial DNA using the PCR method, as reported by Pan and Jiang (Citation2018). The complete mitochondrial genome of P. vachelli is 16,532 bp (MK134689).
The genome of P. vachelli is encoded on two strands and in two directions. The genome contains a total of 37 genes, including 13 protein-coding genes, 22 transfer RNAs, and 2 ribosomal RNAs. The 13 protein-coding genes in the mt genome of P. vachelli also share features in the start and stop codons with those in other species of Siluriformes (Wang et al. Citation2011; Liang et al. Citation2012).
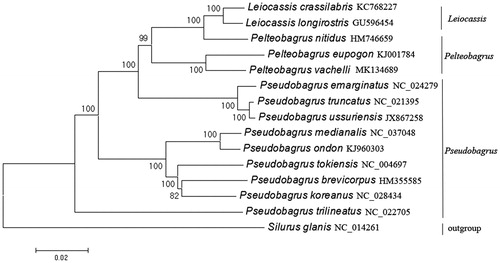
Phylogenetic analysis was performed using 13 protein-coding genes by the neighbor-joining (NJ) method. The clade containing Leiocassis, Pelteobagrus, and Pseudobagrus is well revealed with high supporting value in the phylogenetic tree. It is indicated that Leiocassis and Pelteobagrus are closely related, forming a clade, with Pseudobagrus as a sister group ().
Figure 1. Neighbor-Joining phylogenetic tree inferred from nucleotide sequence dataset of 13 protein-coding genes for 18 Siluriformes mitochondrial genomes. The tree shows the topology based on concatenated data of 13 mitochondrial encoded protein sequences. Reconstruction was performed by MEGA X (64-bit).
Acknowledgments
We thank Dr. Maocang Yan for his comments on the manuscript.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Liang HW, Cao L, Li Z, Zou GW, Liu XL. 2012. Mitochondrial genome sequence of the shining catfish (Pelteobagrus nitidus). Mitochondrial DNA. 23:280–281.
- Pan TS, Jiang H. 2018. The complete mitochondrial genome of Hebesoma violentum (Acanthocephala). Mitochondrial DNA B. 3:582–583.
- Wang JL, Shen T, Ju JF, Yang G. 2011. The complete mitochondrial genome of the Chinese longsnout catfish Leiocassis longirostris (Siluriformes: Bagridae) and a time-calibrated phylogeny of ostariophysan fishes. Mol Biol Rep. 38:280–281.
