Abstract
Oyama sinensis is a large deciduous shrub endemic to Sichuan Province, China. Here, we assembled and characterized the complete chloroplast genome of O. sinensis using Illumina sequencing data. Our assembled chloroplast genome is 160,127 base pairs (bp) in length, containing a large single-copy (LSC) region of 88,206 bp, a small single-copy (SSC) region of 18,783 bp, and two inverted repeat (IR) regions of 26,569 bp. The chloroplast genome possesses total 131 genes, including 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. In addition, phylogenetic analyses show that O. sinensis is closely related to Yulania liliiflora.
Oyama sinensis (Rehder & E. H. Wilson) N. H. Xia & C. Y. Wu (Magnoliaceae) is a large deciduous shrub known only from the mountain forests in Sichuan Province, China (Xia et al. Citation2008). This species has attracted horticultural interest due to the flowers, which are fragrant and showy—white, cup-shaped, 8–12 cm in diameter, with prominent crimson stamens. In addition, the bark of O. sinensis is used in traditional Chinese medicine. Currently, O. sinensis is significantly threatened by habitat loss and considered to be one of the plant species with extremely small populations in the wild (Pan et al. Citation2014). The species is listed as Vulnerable (VU) based on the IUCN Red List criteria (http://www.iucnredlist.org/), and under second-class national protection in China (http://www.forestry.gov.cn/yemian/minglu1.htm). To date, little is known about the population genetics of O. sinensis. Therefore, understanding the genomic characteristics of this endangered species is important for developing conservation strategies. Here, we assembled and characterized the complete chloroplast genome of O. sinensis using Illumina sequencing data.
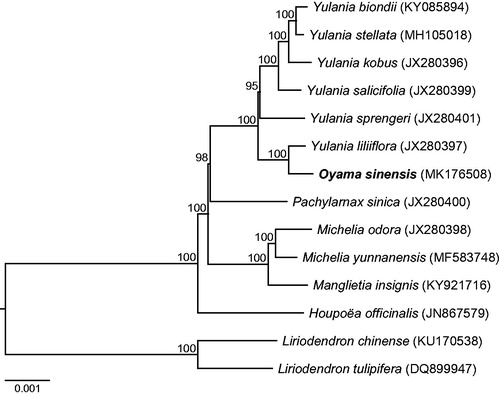
We sampled a single individual of O. sinensis collected from Yingjing County, Sichuan (GPS coordinates: 29°39′40.7″N, 102°36′46.4″E). A voucher specimen (ZhangLS-20180608) was deposited in the Sichuan University Herbarium (SZ). The total genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987). The filtered reads were assembled by the program NOVOPlasty (Dierckxsens et al. Citation2017) for O. sinensis using the sequenced chloroplast genome of its close relative Manglietia insignis (Wallich) Blume (Zheng and Xu Citation2018) as the reference. Next, the assembled chloroplast genome was annotated using Geneious (Kearse et al. Citation2012), and the physical map was generated using OGDRAW (Lohse et al. Citation2013). The chloroplast genome together with gene annotations was submitted to GenBank under the accession number of MK176508. To further investigate its phylogenetic placement, a neighbour-joining tree (Saitou and Nei Citation1987) was constructed by including 13 additional Magnoliaceae species. Here, DNA sequences from these 14 chloroplast genomes were first aligned using MAFFT (Katoh and Standley Citation2013), and a phylogenetic tree was then constructed using MEGA7 (Kumar et al. Citation2016) with 1000 bootstrap replicates.
The chloroplast genome of O. sinensis is 160,127 base pairs (bp) in size, containing a large single-copy (LSC) region of 88,206 bp, a small single-copy (SSC) region of 18,783 bp, and two inverted repeat (IR) regions of 26,569 bp. The chloroplast genome possesses total 131 genes, including 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Most of these genes are in a single copy, however, four rRNA genes (4.5S, 5S, 16S, and 23S rRNA), seven protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, ycf2, and ycf15), and seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC) occur in double copies. The overall GC-content of the whole chloroplast genome is 39.2%, while the corresponding values of the LSC, SSC, and IR regions are 37.9%, 34.3%, and 43.2%, respectively. In addition, phylogenetic analyses show that O. sinensis is closely related to Yulania liliiflora with a 100% bootstrap support ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Pan H-L, Feng Q-H, Long T-L, He F, Liu X-L. 2014. Discussion on resource condition and protection technique for rare endangered species in Sichuan Province. J Sichuan Forest Sci Technol. 35:41–46.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Xia N, Liu Y, Nooteboom HP. 2008. Oyama. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol.7. Beijing, China: Science Press/St. Louis (MO), USA: Missouri Botanical Garden Press; p. 66–68.
- Zheng W, Xu X. 2018. The complete chloroplast genome of endangered Manglietia insignis, a rare landscaping tree with red lotus-like flowers. Conserv Genet Resour. 10:27–30.