Abstract
In this study, we reportted the complete mitochondrial genome of Otocolobus manul. The circular genome was 16,672 bp in length, containing 13 protein coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 non-coding control region. The overall nucleotide composition was as follows: A, 33.1%; T, 27.8%; C, 25.6%; and G, 13.5%, with a total A + T content of 60.9%. The phylogenetic analysis demonstrated a close relationship between O. manul and species in Felis.
The Pallas’ cat (Otocolobus manul), a small-sized felid species that is endemic to Central Asia, is classified as the sole representative of its genus (Otocolobus) belonging to the family Felidae and is noted for its long fur, stocky build, and flattened face (Nowell and Jackson Citation1996; Wang Citation2003). Pallas’ cats are listed as Near Threatened (NT) on the IUCN Red List of Threatened Species and are considered to be threatened with extinction in their natural habitat, primarily because of habitat loss, vermin control programs, and hunting for the fur trade (IUCN 2016). Some researchers argue that the Pallas’ cat should be renamed as Felis manul (Wilson and Reeder Citation2005; Smith et al. Citation2010) and few studies have investigated the molecular taxonomy of wild Pallas’ cats. To clarify the phylogeny and taxonomy of the O. manul, the mitogenome of O. manul was investigated.
The bone sample of O. manul was obtained from the specimen (No. 0038) stored in the mammals’ specimen room of Heilongjiang Province Museum, which was collected from Inner Mongolia Autonomous region (Coordinate: 49.314893 N, 117.737379E), China. The total genomic DNA was extracted from the bone of the specimen using Tissue DNA Extract kit (Tiangen Biotechnique Inc., Beijing, China) following the manufacturer’s protocol stored in Northeast Agricultural University. The mitogenomes were amplified using 20 pairs of primers and PCR products were determined by the method of Sanger sequencing. The sequencing results were then assembled using the DNAman software package (Version 6.0.3, Lynnon Biosoft , Quebec, Canada). The protein-coding genes (PCGs), transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes of the mitochondrial genomes were annotated using the MITOS Web server (Bernt et al. Citation2013).
The mitochondrial genome of O. manul (gb: MH978908) was 16,672 bp in length containing 13 protein-coding, 22 tRNA, and 2 rRNA genes, and 1 non-coding control region. The nucleotide base composition was as follows: A, 33.1%; T, 27.8%; C, 25.6%; and G, 13.5%, with a total A + T content of 60.9%, which was similar to Felis catus, Felis silvestris, and other tortoises in Felinae.
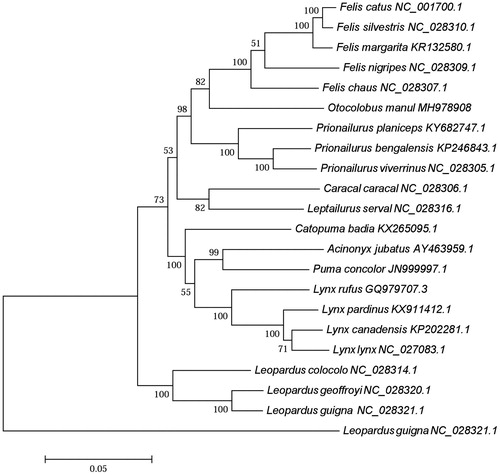
To validate the phylogenetic position of O. manul, a maximum likelihood tree was constructed with MEGA 5.0 (Koichiro, Tokyo, Japan) based on 21 complete mitochondrial genome sequences of Felinae with Leopardus guigna as an outgroup (). Phylogenetic analysis suggested that O. manul was most closely related to Felis catus, Felis silvestris, Felis margarita, Felis nigripes, and Felis chaus, which was supported by 100% bootstrap under ML tree ().
Acknowledgments
The authors thank anonymous reviewers for providing valuable comments on the manuscript.
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogent Evol. 69:313–319.
- Nowell K and Jackson P. 1996. Wild cats: status survey and conservation action plan. IUCN/SSC Cat Specialist Group, Gland, Switzerland. 97–99. Otocolobus manul. [accessed 2018 Sep 27]. http://dx.doi.org/10.2305/IUCN.UK.2016-1.RLTS.T15640A87840229.en
- Smith AT, Gemma F, Xie Y. 2010. A guide to the mammals of china. Changsha: Hunan Education Press.
- Wang YX. 2003. A complete checklist of mammal species and sub-species in China: a taxonomic and geographic reference. Beijing: China Forestry Publishing House.
- Wilson DE, Reeder DM, eds. 2005. Mammal species of the world: a taxonomic and geographic reference. Balitmore (MD): JHU Press.