Abstract
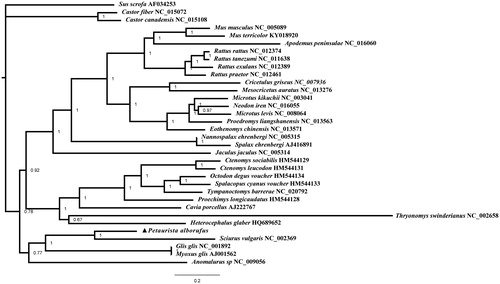
The complete mitochondrial genome of Petaurista alborufus is 16,057 bp in length and contains 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and one control region. In the control region, it is speculated that the CSB2 have been lost in the evolution. The molecular phylogenetic analysis of 32 Rodentia species is performed using sequences from 12 concatenated heavy-strand encoded protein coding genes. The results have provided more evidence to support previous morphological and chromosomal studies on Rodentia.
The Petaurista alborufus, found in southern China, Taiwan, Burma, and Thailand, belongs to the genus Petaurista and is widely distributed throughout South and Southeast Asia (Solari and Baker Citation2006; Oshida et al. Citation2010). In this study, we have determined the complete sequence mitochondrial genome of P. alborufus. This study will be valuable in elucidating the relationship between Pteromyinae and Primates in the future.
The specimen of the wild Petaurista alborufus was obtained from Beichuan, China (N31°56′, E104°16′). The specimen now is stored at the Zoology laboratory at Sichuan Agricultural University. The initial segments of mitochondrial genome were amplified by using LA-PCR technique. We created a master dataset which omitted the ND6 gene and used it for the phylogenetic analysis including only 12 concatenated H-strand protein-coding gene sequences.
The complete mitochondrial genome of Petaurista alborufus is 16,507 bp in size, including 13 protein-coding genes, 22 tRNA genes, two rRNA genes (12S rRNA, 16S rRNA), and one control region (GenBank accession number: JQ743657). We have failed to determine the CSB2, which may have been lost in the evolution. Phylogenetic trees have been obtained by MrBayes (). The complete mitochondrial genome of this throughout South and Southeast Asia and in southern China, Taiwan, and Japan provides a singular novel evolutionary branch to primate taxonomy. Since the genus Petaurista is distributed throughout this area, it can be used as a model genus to test biodiversity and evaluate the evolutionary process.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Oshida T, Dang CN, Nguyen ST, Nguyen NX, Endo H, Kimura J, Sasaki M, Hayashida A, Ai T, Hayashi Y. 2010. Phylogenetics of Petaurista in light of specimens collected from Northern Vietnam. Mammal Study. 35:85–91.
- Solari S, Baker RJ. 2006. Mammal species of the world: a taxonomic and geographic reference. Mastozool Neotrop. 13:290–293.