Abstract
Golden wild yak is a rare and unique Yak subspecies in China. In this study, we first identified the complete mitochondrial genome sequence of golden wild yak. The complete mitogenome was 16,323 bp in length, with 37 typical animal mitochondrial genes (22 transfer RNA, 2 ribosomal RNA and 13 protein-coding genes) and one non-coding region (D-loop), and its contents of A, T, G, and C were 33.71%, 27.26%, 13.21%, 25.82, respectively. The phylogenetic analysis showed that the golden wild yak is closer with Pali yak, and it might be a mutant type of Yak living in Qinghai–Tibetan Plateau area.
Wild yak has high-altitude adaptations such as a strong body (Wiener et al. Citation2003; Dolt et al. Citation2007; Shao et al. Citation2010; Wang et al. Citation2011; Qiu et al. Citation2012), and their hair usually is black, dark brown, tan or white. However, a herd of golden wild yaks whose body is covered with golden hair was found in the Ali region, and the number of golden wild yak was less than 200 in 2015 (Zhou et al. Citation2015). Zhou et al (Citation2015) revealed that golden wild yak is the closest relative to other black wild yaks by their sequence of Cyt b and D-loop (Zhou et al. Citation2015). Until now, the complete mitochondrial genome sequence of golden wild yak has not been identified. Our study aimed to identify the complete mitochondrial genome sequence of the golden wild yak and reveal the phylogenetic status of golden wild Yak in all yak subspecies.
The captive golden wild yak was from Ali Tibetan nature reserve (78 ∼ 86°N and 29 ∼ 35°E). Ear tissues were collected from the captive animals, and Mitochondrial DNA (mtDNA) was extracted. Mitogenome sequence was amplified with 11 primer pairs. PCR products were purified and sequenced by Sanger Sequencing (TSINGKE, Beijing, China). The obtained sequences were assembled using Seqman software. The tRNAs, rRNAs, and protein-coding genes (PCGs) were identified using the tRNAscan-SE 2.0 (Lowe and Eddy Citation1997; Lowe and Chan Citation2016) and online MITOS software (Bernt et al. Citation2013). The neighbor joint (NJ) phylogenetic tree of various cattle species and yak subspecies was constructed using MEGA 7.0 (Wang et al. Citation2010; Liang et al. Citation2016).
The complete mitogenome of Golden wild yak (GenBank No. MK033130) was 16,323 bp in length contain 22 tRNAs, 2 rRNAs, 13 PCGs, and a D-loop. The mtDNA sequence contained 60.97% A + T and 39.03% G + C. We found that nad2, nad3, and nad5 initiated with ATA, whereas the other PCGs started with ATG. These genes have four types of termination codon, including eight PCGs ending with TAA. The terminate codons of nad2 and cob were TAG and AGA, respectively. Another three genes (COX3, ND3, and ND4) ended with the incomplete stop codon ‘T–’, which was presumably formed by the TAA stop codon by post-transcriptional polyadenylation (Anderson et al. Citation1981).
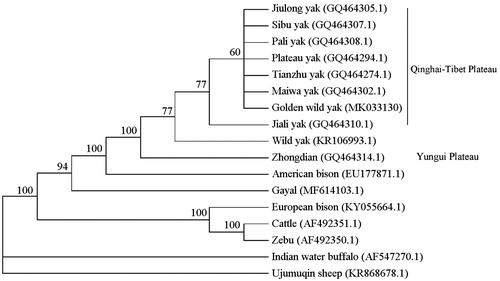
The mitogenome of Golden wild yak was used for taxonomic status analysis combined with other 6 bovine species (American bison, Gayal, European bison, Cattle, Zebu, Water buffalo) and for phylogenetic analysis with other 9 yak subspecies. As shown in , the results showed the taxonomic status of yak (wild yak, golden wild yak and domestic) is close to American bison, and in phylogenetic relationship of yak subspecies, Tianzu, Sibu, Plateau, Maiwa, Pali, Golden, Jiulong, Jiali yak (living in Qinghai–Tibet plateau) were clustered into one branch, and wild yak and Zhongdian yak (living in Yungui plateau) were clustered individually into two branches. The results suggested Golden wild yak might be a mutant type of yak living in Qinghai–Tibetan Plateau area.
Disclosure statement
The authors report no conflict of interest.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290:457–465.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Dolt KS, Mishra MK, Karar J, Baig MA, Ahmed Z, Pasha MA. 2007. cDNA cloning, gene organization and variant specific expression of HIF-1 alpha in high altitude yak (Bos grunniens). Gene. 386:73–80.
- Liang CNA, Wu XY, Ding XZ, Wang HB, Guo X, Chu M, Bao PJ, Yan P. 2016. Characterization of the complete mitochondrial genome sequence of wild yak (Bos mutus). Mitochondrial DNA A. 27:4266–4267.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. 2012. The yak genome and adaptation to life at high altitude. Nature Genetics. 44:946–949.
- Shao B, Long R, Ding Y, Wang J, Ding L, Wang H. 2010. Morphological adaptations of yak (Bos grunniens) tongue to the foraging environment of the Qinghai-Tibetan Plateau. J Animal Sci. 88:2594.
- Wang HC, Long RJ, Liang J, Guo XS, Ding LM, Shang ZH. 2011. Comparison of nitrogen metabolism in yak (Bos grunniens) and indigenous cattle (Bos taurus) on the Qinghai-Tibetan Plateau. Asian-Australasian J Animal Sci. 24. 766–773.
- Wang Z, Shen X, Liu B, Su J, Yonezawa T, Yu Y, Guo S, Ho SYW, Vilà C, Hasegawa M, et al. 2010. Phylogeographical analyses of domestic and wild yaks based on mitochondrial DNA: new data and reappraisal. J Biogeo. 37:2332.
- Wiener G, Han JL, Long RJ. 2003. The yak. Bangkok, Thailand: RAP Publication. pp. 57–58.
- Zhou YY, Zhang YG, Lu H, Liu F, Li DQ, Feng JC. 2015. Preliminary analysis on taxonomic status of golden wild yak in Tibet. Acta Theriologica Sinica. 35:48–54.