Abstract
Allium monanthum is a typical spring ephemeral, appearing above ground for only 2–3 weeks in late March to early or late April. Here, Illumina pair-end reads of this species was used to record the complete chloroplast (cp) genome. The circular genome with 154,804 bp in length contains a pair of inverted repeats (IRs) of 24,551 bp, separated by a large single-copy region (LSC) of 83,701 bp and a small single-copy region (SSC) of 21,867 bp. It harbors 132 genes, including 86 protein-coding genes, 38 transfer RNA genes, and 8 ribosomal RNA genes. Phylogenetic analysis supported that the A. monanthum was located in the crown of the Alliaceae, and was closely clustered with other Allium species.
A typical spring ephemeral, Allium monanthum Maxim., which belongs to the section Microscordum of the genus Allium (Alliaceae) (Traub Citation1968), appears above ground for only 2–3 weeks from late March to early or late April (Kawano et al. Citation2005). This species possesses very peculiar life history features, including complex sexuality and characteristic vegetative reproduction (Kawano Citation1968; Nagai Citation1972; Kawano and Nagai Citation1975). The tiny wild onion species is a typical monocarpic ‘pseudo-annual’, having one or two (or rarely, three) small aerial leaves measuring only 10 cm long and 4–6 mm wide, and small bulbs that are entirely renewed each season (Kawano et al. Citation2005). In addition, it has been used as an important germplasm resource for genetic breeding of wild onion and is well-known for its nutritional (Kunkel Citation1984) and material uses (Riotte Citation1978; Zhong and Huang Citation1980). Here, we reported the complete chloroplast (cp) genome of A. monanthum, and deposited the annotated cp genome into GenBank with the accession number MH748538.
Fresh leaves were collected from Jiamusi (Heilongjiang province, China; coordinates: 46°33′46.46″N, 104°36′6.96″E), and total genomic DNA was extracted from leaf tissues with the modified CTAB method (Doyle and Doyle Citation1987). Voucher specimens were deposited in the Herbarium of Sichuan University (SZ). The whole genome sequencing was achieved on the Illumina Hiseq Platform (Illumina, San Diego, CA). The complete chloroplasts genome was assembled via NOVOPlasty (Dierckxsens et al. Citation2017) with the sequences of published Allium victorialis chloroplast genome as reference (GenBank accession number is NC_037240). We annotated the plastid genome using Plann v1.1 (Huang and Cronk Citation2015) and corrected the annotation with Geneious v11.0.3 (Kearse et al. Citation2012). Finally, the genome map was generated by using the web server OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) (Lohse et al. Citation2013).
The complete cp genome of A. monanthum was 154,804 bp in length, containing a large codon (LSC, 83,701 bp) and a small codon (SSC, 21,867 bp) single-copy regions, separated by a pair of inverted repeat regions (IRs, 24,551 bp each). The genome harbors 132 genes, including 86 protein-coding genes (PCG), 38 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). Most of the genes occur as a single copy in LSC or SSC, while 18 genes are duplicated in the IR regions, including seven PCG species, seven tRNA, and four rRNA species. The overall GC content of A. monanthum cp genome was 37.0%, with the corresponding values of LSC, SSC, and IR regions being 34.8%, 31.3%, and 43.2%, respectively.
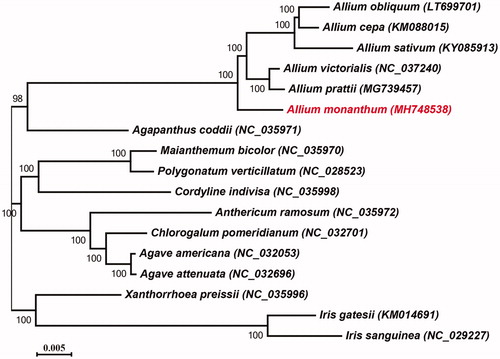
To ascertain the phylogenetic relationship between A. monanthum and other Allium taxa, 16 cp genome sequences of nine genera were achieved from the National Center for Biotechnology Information (NCBI, https://www. ncbi.nlm.nih.gov/). After aligning by MAFFT, the alignment was used to construct a maximum likelihood (ML) tree via RAxML v8 (Stamatakis Citation2014) with 200 bootstrap replicates (). The result showed that A. monanthum was located in the stem of the Alliaceae, and closely clustered with other Allium species, which was in agreement with with the previous studies (Friesen et al. Citation2006; Li et al. Citation2010). The complete chloroplast genome data provide fundamental information for the recongnition and utilization for A. monanthum. This genome will also contribute significantly to the phylogenetic and evolutionary studies of the family Alliaceae.
Disclosure statement
The authors are grateful to the opened raw genome data from public database, and the authors declare no conflicts of interest and are responsible for the content.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Friesen N, Fritsch RM, Blattner FR. 2006. Phylogeny and new intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso. 22:372–395.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kawano S. 1968. Problems in experimental taxonomy, with special reference to reproductive and productive biology of the flowering plants. Proc Jpn Soc Plant Taxon. 2:23–29.
- Kawano S, Nagai Y. 1975. The productive and reproductive biology of flowering plants I. Life history strategies of three Allium species in Japan. Botan Mag Tokyo. 89:281–318.
- Kawano S, Nagai Y, Hayashi K. 2005. Life‐history monographs of Japanese plants. 3: Allium monanthum maxim. (Alliaceae). Plant Spec Biol. 20:155–165.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kunkel Gnther. 1984. Plants for human consumption. Kew Bull. 42:273.
- Li QQ, Zhou SD, He XJ, Yu Y, Zhang YC, Wei XQ. 2010. Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann Bot London. 106:709.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Nagai Y. 1972. Reproductive biology of Allium monanthum Maxim. J Geobot. 20:84–91.
- Riotte L. 1978. Companion planting for successful gardening. Vermont (USA): Garden Way.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Traub HP. 1968. The subgenera, sections and subsection of Allium L. Plant Life. 24:147–163.
- Zhong XT, Huang XT. 1980. Flora of China. 14:267.