Abstract
Mouthbrooding fighting fish Betta apollon and B. simplex are widely distributed in Southeast Asia but urbanization is restricting their biodiversity. Complete mitochondrial genomes (mitogenomes) of B. apollon and B. simplex were determined to support systematic conservation programs. Mitogenome sequences were 16,536 and 16,549 bp in length with slight AT bias (56.68% and 56.60%), respectively, containing 37 genes with the order identical to most teleost mitogenomes. Phylogenetic analysis of B. apollon showed a closer relationship with B. simplex, grouped with B. pi as a monophyletic clade of mouthbrooders. Results will facilitate evolutionary studies, species diversity, and conservation management in fighting fishes.
Fighting fish of the genus Betta in Southeast Asia are separated into two groups as mouthbrooding and bubble nesting (Ruber et al. Citation2004). Comparison of these two fighting fish groups indicates that bubble nesting males are more colorful and easier to identify than mouthbrooder males (Panijpan et al. Citation2014). However, morphological observations show that larval states in both fighting fish groups are very similar at the species level. This results in operational difficulties for farming conservation programs, and biodiversity is decreasing globally due to urbanization. Study of genetic diversity in fighting fishes is, therefore, required to provide information for prospective breeding and conservation management. Here, complete mitochondrial genomes (mitogenomes) of B. apollon and B. simplex collected from Narathiwat Province (6.1789N, 102.0547E) (No. THNHN-F001551) and Krabi Province (7.9307 N, 99.2552 E) (No. THNHN-F001552), respectively, were determined and stored in the Thailand Natural History Museum. Whole genomic DNA was extracted in accordance with the standard salting-out protocol (Supikamolseni et al. Citation2015). Polymerase chain reaction (PCR) amplifications were performed using universal mitogenome primers (Mauro et al. Citation2004), and specific primers for both Betta species were developed to amplify remaining parts of the genome. All PCR products were sequenced by the DNA sequencing service of First Base Laboratories Sdn Bhd (Selangor, Malaysia), with annotation performed following Srikulnath et al. (Citation2012) and Prakhongcheep et al. (Citation2018).
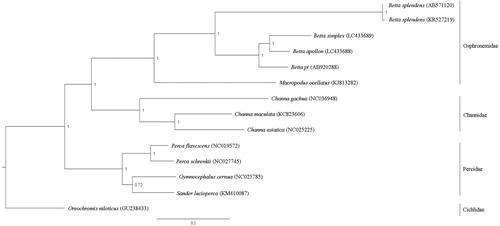
Complete mitogenome sequences consisted of 16,536 bp for B. apollon (GenBank accession no. LC433688) and 16,549 bp for B. simplex (LC433689). Both mitogenomes contained 37 genes and a control region (CR). Gene arrangement patterns were identical to those of teleosts (Miya et al. Citation2013). Overall AT content values for the mitochondrial genome were 56.68% for B. apollon and 56.60% for B. simplex. Nucleotide diversity (p-distance) among all Betta mitogenomes was determined at 15.54 ± 0.03%, and among mouthbrooders at 10.82 ± 0.05%. Four conserved sequence blocks (CSB-D, CSB1, CSB2, and CSB3) in the CR of teleost mitogenomes were also present in B. apollon and B. simplex (Lee and Kocher Citation1995; Prakhongcheep et al. Citation2018). No tandem repeat was found in the CR of B. apollon; however, a tandem repeat was identified in B. simplex at 15,707–15,749 bp. Diverse numbers of tandem repeats were observed in B. pi (AB920288) and B. splendens (AB571120 and KR527219) (Song et al. Citation2016; Prakhongcheep et al. Citation2018), suggesting that the CR had a large variation in different fighting fish species. A phylogenetic tree was constructed based on 12 concatenated protein-coding genes without ND6 of 14 teleosts, using Bayesian inference with MrBayes version 3.2.6 (Huelsenbeck and Ronquist Citation2001). The sister group comprising B. apollon and B. simplex formed a monophyletic clade with B. pi as mouthbrooders, consistent with Panijpan et al. (Citation2014). The fighting fish clade was also close to the Macropodus clade, confirming results from previous studies (Ruber et al. Citation2004) (). These complete mitogenomes will enrich genomic resources for future evolutionary and diversity studies to improve conservation management.
Figure 1. Phylogenetic relationships among twelve concatenated mitochondrial protein-coding genes, without ND6 sequences of fourteen mitochondrial genomes including Oreochromis niloticus as the outgroup using Bayesian inference analysis. The complete mitochondrial genome sequence was downloaded from GenBank. Accession numbers are indicated in parentheses after the scientific names of each species. Support values at each node are Bayesian posterior probabilities while branch lengths represent the number of nucleotide substitutions per site.
Acknowledgements
The authors would like to thank Sunchai Makchai (Thailand Natural History Museum) for advice on sample preparation.
Disclosure statement
The authors report no conflicts of interest and are entirely responsible for the contents of this article. Animal care and all experimental procedures were approved by the Animal Experiment Committee, Kasetsart University, Thailand (approval no. ACKU01157) and conducted in accordance with the Regulations on Animal Experiments at Kasetsart University.
Additional information
Funding
References
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Lee WJ, Kocher TD. 1995. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome: early establishment of the vertebrate genome organization. Genetics. 139:873–887.
- Mauro DS, David JJG, Oommen VO, Wilkinson M, Zardoya R. 2004. Phylogeny of caecilian amphibians (Gymnophiona) based on complete mitochondrial genomes and nuclear RAG1. Mol Phylogenet Evol. 33:413–427.
- Miya M, Friedman M, Satoh TP, Takeshima H, Sado T, Iwasaki W, Yamanoue Y, Nakatani M, Nakatani K, Inoue JG, et al. 2013. Evolutionary origin of the Scombridae (Tunas and Mackerels): members of a paleogene adaptive radiation with 14 other pelagic fish families. PLoS One. 8:e73535.
- Panijpan B, Kowasupat C, Laosinchai P, Ruenwongsa P, Phongdara A, Senapin S, Wanna W, Phiwsaiya K, Kühne J, Fasquel F. 2014. Southeast Asian mouth-brooding Betta fighting fish (Teleostei: Perciformes) species and their phylogenetic relationships based on mitochondrial COI and nuclear ITS1 DNA sequences and analyses. Meta Gene. 2:862–879.
- Prakhongcheep O, Muangmai N, Peyachoknagul S, Srikulnath K. 2018. Complete mitochondrial genome of mouthbrooding fighting fish (Betta pi) compared with bubble nesting fighting fish (B. splendens). Mitochondrial DNA B. 3:6–8.
- Ruber L, Britz R, Tan HH, Ng PKL, Zardoya R. 2004. Evolution of mouthbrooding and life-history correlates in the fighting fish genus Betta. Evolution. 58:799–813.
- Song YN, Xiao GB, Li JT. 2016. Complete mitochondrial genome of the Siamese fighting fish (Betta splendens). Mitochondrial DNA A. 27:4580–4581.
- Srikulnath K, Thongpan A, Suputtitada S, Apisitwanich S. 2012. New haplotype of the complete mitochondrial genome of Crocodylus siamensis and its species-specific DNA markers: distinguishing C. siamensis from C. porosus in Thailand. Mol Biol Rep. 39:4709–4717.
- Supikamolseni A, Ngaoburanawit N, Sumontha M, Chanhome L, Suntrarachun S, Peyachoknagul S, Srikulnath K. 2015. Molecular barcoding of venomous snakes and species-specific multiplex PCR assay to identify snake groups for which antivenom is available in Thailand. Genet Mol Res. 14:13981–13997.
