Abstract
Phoebe zhennan S. Lee is the most important Phoebe plant, which is also a rare and extremely valuable wood in China. In this study, we obtained the complete chloroplast genome sequence of P. zhennan S. Lee using BGISEQ-500 sequencing. Its chloroplast genome size has 152,212 bp length, containing a large single-copy region (93,755 bp), a small single-copy region (18,929 bp), and a pair of IR regions (19,764 bp). The overall GC contents of the chloroplast genome were 39.9%. This circular genome contains 127 annotated genes, including 81 protein-coding genes, 38 tRNAs, and 8 rRNAs. The phylogenetic analysis using maximum likelihood (ML) method showed that P. zhennan S. Lee has the closest relationship with P. nanmu. This complete chloroplast genomes can be subsequently used for the forestry development of this valuable species.
Phoebe zhennan S. Lee belongs to the family Lauraceae and the main tree species of evergreen broad-leaved forest. It is a precious timber and landscape tree species, endemic to China, where it is listed as a nationally protected vulnerable species (Gao et al. Citation2016). The existing natural population of P. zhennan S. Lee is scattered in the Sichuan, Guizhou, Hubei, and Hunan below 1500 M above sea level, and the wild resources are gradually reducing now (Hu et al. Citation2015). In order to further study the genetic diversity and genetic structure of natural population in P. zhennan S. Lee, it can be considered as endangered species to retain as much genetic variation as possible to enhance the chance for its recovery. However, our understanding of the origins of the chloroplast genome of this forestry plant is limited. In this study, we obtained the complete chloroplast genome of P. zhennan S. Lee and explored the phylogenetic relationship with other species, which contributes to phylogenetic studies of these taxa and the sustainable forestry development of this valuable species.
The specimen of P. zhennan S. Lee was isolated from Longtan Village Dahe Town test field in Enshi, Hubei, China (109.09E; 29.50N). The total DNA of P. zhennan S. Lee was extracted using the CTAB method and stored in Enshi Tujia and Miao Autonomous Prefecture Academy of Forestry (No. ETMAPAF01). The DNA sample was sequenced using the BGISEQ-500 Sequencing Platform (BIG, Shenzhen, CA, CHN). Quality control was performed to remove low-quality reads and adapters using the FastQC software (Andrews Citation2015). The chloroplast genome was assembled with SPAdes v3.8 (Bankevich et al. Citation2012) and annotated by Blast and DOGMA (Wyman et al. Citation2004). The tRNA genes were further identified using tRNAscan-SE 2.0 (Lowe and Chan Citation2016). The annotated chloroplast genome was submitted to the GenBank database under accession No. MH158313.
The complete chloroplast genome of P. zhennan S. Lee was a circle with 152,212 bp in size, containing a large single-copy region (LSC) of 93,755 bp, a small single-copy region (SSC) of 18,929 bp, and a pair of inverted repeat regions (IRs) of 19,764 bp. In total, 127 genes were annotated on this chloroplast genome, including 81 protein-coding genes (PCG), 38 transfer RNA genes (tRNA), and 8 ribosomal RNA genes (rRNA). In the IR regions, a total of 12 genes were found duplicated, including 2 PCG species (rps7 and rps12), 6 tRNA species (trnL-CAA, trnV-GAC, trnI-CAU, trnA-UGC, trnR-ACG, and trnN-GUU) and 4 rRNA species (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition is: 30.0% A, 30.8% T, 19.9% C, and 20.0% G, with a total G + C content of 39.9%.
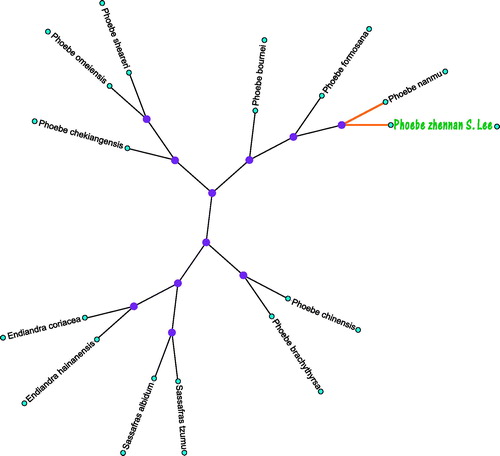
For phylogenetic analysis, we selected other 12 plants chloroplast genomes from GenBank to assess the relationship of P. zhennan S. Lee. The genome-wide alignment of all chloroplast genomes was constructed by HomBlocks(Bi et al. Citation2018). The phylogenetic trees were reconstructed using maximum likelihood (ML) methods. ML analysis was performed using RaxML-8.2.4(Stamatakis Citation2014), of which the bootstrap values were calculated using 5000 replicates to assess node support and all the nodes were inferred with strong support by the ML methods. The final tree was represented and edited using MEGA X (Kumar et al. Citation2018). As shown in the phylogenetic tree (), the chloroplast genome of P. zhennan S. Lee was clustered with P. nanmu.
Figure 1. The maximum likelihood (ML) tree inferred from P. zhennan S. Lee and other 12 plants chloroplast genomes. This tree was drawn without setting out groups. All nodes exhibit above 90% bootstraps. The length of branch represents the divergence distance. The NCBI database accession number of P. zhennan S. Lee to P. zhennan S. Lee in the counterclockwise direction is, NC036143.1, NC272328.1, NC034926.1, NC031191.1, NC031190.1, NC034925.1, HM268714.1, HM268814.1, AF268832.1, AF272336.1, NC0372339.1, NC038324.1.
Acknowledgments
This research was supported by Science and technology plan projects of Enshi (No. D20180010);Science and Technology Promotion Project of Forestry from Central Finance (No. 2018TG01).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Gao J, Zhang W, Li J, Long H, He W, Li X. 2016. Amplified fragment length polymorphism analysis of the population structure and genetic diversity of Phoebe zhennan (Lauraceae), a native species to China. Biochem Systematics Ecol. 64:149–155.
- Hu Y, Wang B, Hu T, Chen H, Li H, Zhang W, Zhong Y, Hu H. 2015. Combined action of an antioxidant defence system and osmolytes on drought tolerance and post-drought recovery of Phoebe zhennan S. Lee saplings. Acta Physiol Plantarum. 37:84.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lowe, TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
