Abstract
Rhynchocypris oxycephalus (Sauvage and Dabry de Thiersant, 1874) distributed in East Asia is small cold-water fish. We completed whole mitochondrial genome of R. oxycephalus caught in Korea. Its length is 16,583 bp, which is shorter than those of three mitochondrial genomes from China and Japan. Three hundred ninety seven of 1,278 SNPs and 27 of 44 INDELs are Korean R. oxycephalus specific. It contains 13 protein-coding genes, 2 rRNAs and 22 tRNAs and its GC ratio is 44.5%. Phylogenetic analysis presents genetic diversity of R. oxycephalus originated from Korea, Japan, and China.
The Rhynchocypris minnows, covering seven species, are small cold-water fish, mainly distributed in Asia (Chen Citation1998). Rhynchocypris oxycephalus is distributed only in East Asia covering southern China, Korea, and Eastern Japan (Chen Citation1998; Park et al. Citation2001; Ito et al. Citation2002; Sakai et al. Citation2006; Bogutskaya et al. Citation2008). Chinese minnow usually lives in mountain stream in Korea because it prefers fresh water. It is an indicator species of first-grade fresh water. Its morphology is similar to that of amur minnow (Rhynchocypris lagowskii) and both species have same mitochondrial DNA haplotype caused by introgression of mitochondrial DNA via interspecific hybridization (Sakai et al. Citation2006). To understand phylogenetic relationship of R. oxycephalus in different countries with three mitochondrial genomes from China (Sui et al. Citation2016) and Japan (Imoto et al. Citation2013), we sequenced mitochondrial genome of R. oxycephalus in Korea.
Chinese minnow was caught in Mt. Suri, Gyounggi province, Korea (Voucher in InfoBoss Cyber Herbarium (IN); IBS-00009). Total DNA was extracted from fresh flesh of R. oxycephalus by using a DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008). All bases were confirmed by alignment results generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd, Auckland, New Zealand) was used for mitochondrial genome annotation based on R. oxycephalus mitochondrial genome (NC_018818).
The mitochondrial genome of R. oxycephalus (Genbank accession is MK208924) is 16,583 bp, which is shorter than those of R. oxycephalus in China (by 23 bp; Sui et al. Citation2016) and Japan (by 25–26 bp; Imoto et al. Citation2013), caused by 28 bp gap in D-loop region of Korean R. oxycephalus mitochondrial genome. GC ratio of the mitochondrial genome is 44.5% and it contains 13 protein-coding genes, two rRNAs, and 22 tRNAs, which is the same as those of 18 available Rhynchocypris mitochondrial genomes.
Based on alignment of four mitochondrial genomes of R. oxycephalus, 1,278 of 16,615 (7.7%) sites contain single nucleotide polymorphisms (SNPs) and 44 sites have insertions and deletions (INDELs). 397 out of 1,278 SNPs (31.1%) and 27 out of 44 INDELs (61.4%) are Korean specific. 27 Korean specific INDELs are from 28 bp gap where Japanese R. oxycephalus (NC_018818) presents only one INDEL.
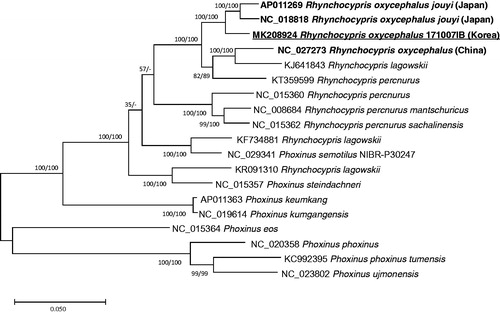
Nineteen complete mitochondrial genomes in Rhynchocypris and Phoxinus genera were analyzed for constructing phylogenetic trees using maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) methods. Multiple sequence alignment was conducted with nineteen complete mitochondrial genomes by MAFFT 7.388 (Katoh and Standley Citation2013) and phylogenetic trees were generated by MEGA X (Kumar et al. Citation2018). Both trees present that R. oxycephalus from Japan and Korea are clustered in one clade, while that of China is in another clade (). Interestingly, three mitochondrial genomes of R. lagowskii are scattered in three clades (), which is different from the expectation derived from similar morphological characters between two species. Altogether, our mitochondrial genome addresses the question of evolutionary history of R. oxycephalus among three countries.
Figure 1. Maximum likelihood and neighbor joining phylogenetic trees of 19 Rhynchocypris and Phoxinus mitochondrial genomes: Rhynchocypris oxycephalus (MK208924 in this study), Rhynchocypris oxycephalus jouyi (AP011269), Rhynchocypris oxycephalus jouyi (NC_018818), Rhynchocypris oxycephalus IHB-LHG20130605 (NC 027273), Rhynchocypris lagowskii (KJ641843), Rhynchocypris percnurus (KT359599), Rhynchocypris percnurus (NC_015360), Rhynchocypris percnurus mantschuricus (NC_008684), Rhynchocypris percnurus sachalinensis (NC_015362), Rhynchocypris lagowskii (KF734881), Rhynchocypris lagowskii (KR091310), Phoxinus semotilus NIBR-P30247 (NC 029341), Phoxinus steindachneri (NC_015357), Phoxinus keumkang (AP011363), Phoxinus kumgangensis (NC_019614), Phoxinus eos (NC_015364), Phoxinus phoxinus (NC_020358), Phoxinus phoxinus tumensis (KC992395), and Phoxinus ujmonensis (NC_023802). The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bogutskaya NG, Naseka AM, Shedko SV, Vasileva ED, Chereschnev I. 2008. The fishes of the Amur River: updated check-list and zoogeography. Ichtyol Explor Freshw. 19:301–366.
- Chen Y. 1998. Fauna Sinica, Osteichthyes, Cypriniformes (middle volume). Beijing: Science Press. (in Chinese).
- Imoto JM, Saitoh K, Sasaki T, Yonezawa T, Adachi J, Kartavtsev YP, Miya M, Nishida M, Hanzawa N. 2013. Phylogeny and biogeography of highly diverged freshwater fish species (Leuciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene. 514:112–124.
- Ito Y, Sakai H, Shedko S, Jeon SR. 2002. Genetic differentiation of the northern Far East cyprinids, Phoxinus and Rhynchocypris. Fisheries Sci. 68:75–78.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Park IS, Im JH, Ryu DK, Nam YK, Kim DS. 2001. Effect of starvation on morphometric changes in Rhynchocypris oxycephalus (Sauvage and Dabry). J Appl Ichthyol. 17:277–281.
- Sakai H, Ito Y, Shedko SV, Safronov SN, Frolov SV, Chereshnev IA, Jeon S-R, Goto A. 2006. Phylogenetic and taxonomic relationships of northern Far Eastern phoxinin minnows, Phoxinus and Rhynchocypris (Pisces, Cyprinidae), as inferred from allozyme and mitochondrial 16S rRNA sequence analyses. Zool Sci. 23:323–331.
- Sui X, Liang Y, He D. 2016. The complete mitochondrial genome of Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae). Mitochondrial DNA. 27:3367–3369.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
