Abstract
The sea potato (Acaudina molpadioides) is a species of sea cucumber of the family Holothuroidea under Phylum Echinodermata. In this study, we firstly sequenced and annotated the complete mitochondrial genome of A. molpadioides, which was 16,028 bps in size and contained 13 protein-coding genes, 2 rRNAs, 19 tRNA genes, and 1 putative control region. The overall A + T content (65.47%) was higher than G + C content (34.53%), showing the AT bias, consistently with other invertebrate mitochondrial genomes. Phylogenetic analyses showed that the mitochondrial DNAs of A. molpadioides were closely related to other species of Holothuroidea.
The sea potato (Acaudina molpadioides) is a species of sea cucumber widely distributed in East Asia counties. Most of this species live in the intertidal zone and only few of them live in the coast sediment. Since A. molpadioides has high fertility, it already has caused serious damage to the marine environment. The existing reports are limited to the study of nutrients and chemical constituents of A. molpadioides (Ping and Yang Citation2013; Yu et al. Citation2013), while the molecular data is largely lacking. Mitochondrial genome data has been utilized to study phylogeographic (Scribner et al. Citation2003), genetic diversity and species identification (Curole and Kocher Citation1999, and phylogenetic relationships of the animals in the metazoans (Boore et al. Citation2005). In this study, we successfully completed the sequencing and annotated the mitochondrial genome of A. molpadioides.
Adults A. molpadioides were collected from the sea surrounding Ningde, Fujian province, China. After morphological identification, the samples were preserved in 75% ethanol and deposited at the Laboratory for Morphogenesis and Evolution, Ocean University of China (Specimen code: OUC-MLS-2018-MIFJ00-01-45). Total DNA was extracted from the gonad using phenol/chloroform/isoamyl alcohol method (Asahida et al. Citation1996) and stored at −20 °C. The complete mitochondrial genome of A. molpadioides was amplified using polymerase chain reaction (PCR) (Cheng et al. Citation1994; Boore et al. Citation2005).
The assembled complete mitochondrial genome of A. molpadioides was 16,028 bps. Genome sequence was submitted to the Gen-bank with accession number MK050109. The average nucleotide composition of the mitochondrial genome was A (34.98%), T (30.49%), C (21.13%), and G (13.40%). Thirteen protein-coding genes, 2 ribosomal RNA genes, 19 transfer RNA genes, and a sequence of putative control region were identified in mitochondrial genome of A. molpadioides. The gene arrangement of A. molpadioides mtDNA was identical to most of Holothurian mitochondrial genomes (Boore Citation1999; Fan et al. Citation2011). The 13 conserved protein-coding genes were cytochrome oxidase subunit (I, II, III), NADH dehydrogenase subunit (1, 2, 3, 4, 5, 6 and 4L), ATP synthase subunit (6, 8), and cytochrome b apoenzyme. Most of the protein-coding genes were encoded on the light strand accepting that NADH dehydrogenase subunit 6 encoded on the heavy strand. The products of the two rRNA were small subunit ribosomal RNA (12SrRNA) and large subunit ribosomal RNA (16SrRNA), respectively. Nineteen tRNA genes were predicted and the typical clover leaf structures formed by nucleotides folding were present in all transfer RNA genes.
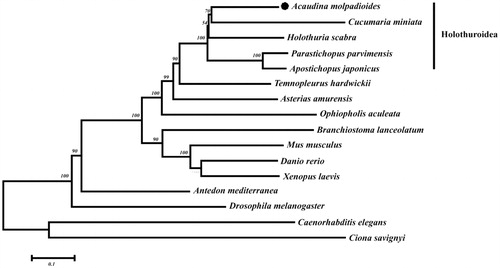
In order to clearly understand the evolutionary status of A. molpadioides, the mitochondrial sequences of A. molpadioides, its closely related species (Apostichopus japonicus, Parastichopus parvimensis, Cucumaria miniata, Holothuria scabra), echinoderms (Ophiopholis aculeata, Antedon mediterranea, Asterias amurensis, Temnopleurus hardwickii), and some model organisms (Caenorhabditis elegans, Drosophila melanogaster, Ciona savignyi, Branchiostoma lanceolatum, Danio rerio, Xenopus laevis, and Mus musculus) were downloaded from NCBI GenBank datebase for phylogenetic relationship analysis. Phylogenetic tree was constructed based on the maximum-likelihood method to show the evolutionary relationships. Phylogenetic tree results showed that A. molpadioides formed a clade with were other species of Holothuroidea (). This result provides strong evidence that A. molpadioides belongs to Holothuroidea family.
Figure 1. Phylogenetic tree of complete mitochondrial genome of A. molpadioides and other species. The accession numbers for all species are as follows:Acaudina molpadioides (MK050109), Apostichopus japonicas (FJ906623.1), Parastichopus parvimensis (KU168761.1), Cucumaria miniata (AY182376.1), Holothuria scabra (KP257577.1), Ophiopholis aculeate (AF314589.1), Antedon mediterranea (NC_010692.1), Asterias amurensis (NC_006665.1), Temnopleurus hardwickii (KP070768.1), Caenorhabditis elegans (NC_001328.1), Drosophila melanogaster (KT896664.1), Ciona savignyi (NC_004570.1), Branchiostoma lanceolatum (NC_001912.1), Danio rerio (NC_002333.2), Xenopus laevis (NC_001573.1), and Mus musculus (KY018919.1).
Disclosure statement
The authors report no competing interests declared. All data is true and valid.
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction form fish stored at ambient temperature using buffers containing high concentration of urea. Fisheries Sci. 62:727–730.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Boore JL, Macey JR, Medina M. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 395:311.
- Cheng S, Chang SY, Gravitt P, Respess R. 1994. Long PCR. Nature. 369:684–685.
- Curole JP, Kocher TD. 1999. Mitogenomics: digging deeper with complete mitochondrial genomes. Trends Ecol Evol. (Amst.). 14:394–398.
- Fan S, Hu C, Wen J, Zhang L. 2011. Characterization of mitochondrial genome of sea cucumber Stichopus horrens: a novel gene arrangement in Holothuroidea. Sci China Life Sci. 54:434–441.
- Ping Y, Yang W. 2013. Extraction of collagen from Acaudina molpadioides and the type determination and optimization of hydrolyzed conditions. J Chinese Inst Food Sci Technol. 13:94–98.
- Scribner KT, Talbot SL, Pearce JM, Pierson BJ, Bollinger KS, Derksen DV. 2003. Phylogeography of Canada geese (Branta canadensis) in western North America. Auk. 120:889–907.
- Yu L, Xu XQ, Xue CH, Chang YG, Ge L, Wang YC, Zhang CY, Liu GC, He C. 2013. Enzymatic preparation and structural determination of oligosaccharides derived from sea cucumber (Acaudina molpadioides) fucoidan. Food Chem. 139:702–709.
