Abstract
Dactylellina cionopaga is a nematophagous fungus known to form unstalked adhesive knobs and simple two-dimension networks. In our study, we sequenced almost complete mitochondrial genome of D. cionopaga. The mitogenome has 194,125 base pairs with a GC content of 26.59% and made up of total of 32 genes (14 protein-coding genes, 16 transfer RNAs genes, and two ribosomal RNAs genes). The complete mitogenome of D. cionopaga will provide potential genetic resources for further evolutionary studies of this species.
Dactylellina cionopaga (Ascomycota, Orbiliomycetes, Orbiliales, Orbiliaceae) is a trapping fungus that was isolated from root-zone soil (Yu, Duan, et al. Citation2012) of the Panax in China (Yunnan). This species capturing nematode by producing unstalked adhesive knobs and a two-dimensional network (Zhang et al. Citation2011), which can trap Caenorhabditis elegans (Yu, Xue, et al. Citation2012) and this connection may provide a model system to understanding the interaction between predacious fungus and plant parasitic nematodes. However, less is known about its mitogenome. In this study, we report the complete mitogenome sequence and the phylogenetic relationships of D. cionopaga in relation to other ascomycetous fungi based on mitochondrial proteins.
The mitogenome was extracted from the whole genome sequence of a pure culture of strain YMF1.00569 (collected from soil, at Deqen County, Diqing Tibetan Autonomous Prefecture, by Lu Cao, 2004. N 29° 15' E 99° 32'). Whole genome sequencing was performed using the MiSeq (Illumina, San Diego, CA). Reads were assembled using A5-miseq 20150522 (Coil et al. Citation2015) and SPAdes 3.9.0 (Bankevich et al. Citation2012). The mitogenome was identified using BLAST with Pyronema omphalodes (GenBank ID: KU707476) as a reference. The complete mitogenome was annotated using MFANNOT(http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl), and transfer RNA (tRNA) genes were identified using tRNAscan-SE1.2.1 (Lowe and Eddy Citation1997) with the default search mode. All ORFs were searched and identified using ORFFinder (https://www.ncbi.nlm.nih.gov/orffinder?tdsourcetag=s_pcqq_aiomsg).
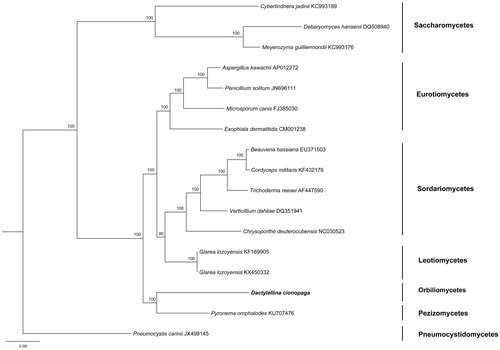
The mtDNA of D. cionopaga consists of a single circular chromosome 194,125 bp in length with a GC content of 22.92%. The mitogenome sequenced harbours 14 protein-coding genes (PCGs), 16 transfer RNA genes (tRNA), and two ribosomal RNA genes (rnl and rns). The 14 conserved protein-coding genes include nad1 (four introns), nad2 (five introns), nad3, nad4 (two introns), nad4L, nad5 (five introns), nad6, cox1 (12 introns), cox2 (two introns), cox3 (five introns), atp6 (three introns), atp8, atp9 (four introns), and cob (six introns). All tRNA genes are encoded on the sense strand. Phylogenetic relationship of D. cionopaga was inferred from 14 common mitochondrial protein-coding genes of other closely related species was performed using Bayesian inference (BI). Pneumocystis carinii derived from Pneumocystidaceae was used as out group for tree rooting. The BI tree () showed that D. cionopaga grouped with Pyronema omphalodes. The Genbank Accession number for Dactylellina cionopaga is MK307796.
Figure 1. Phylogenetic relationships using Bayesian analysis among 17 Ascomycota fungi based on the concatenated amino acid sequences of 14 mitochondrial protein-coding genes. The following 14 mitochondrial protein-coding genes were concatenated: atp6, atp8, atp9, cytb, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6. Pneumocystis carinii was used as out-group. Bayesian posterior probabilities are shown at nodes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31:587–589.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955.
- Yu H, Duan J, Wang B, Jiang X. 2012. The function of snodprot in the cerato-platanin family from Dactylellina cionopaga in nematophagous fungi. Biosci Biotechnol Biochem. 76:1835–1842.
- Yu HY, Xue W, Gao XX. 2012. Development of a transformation method for the nematophagous fungus Dactylellina cionopaga. Afr J Biotechnol. 11:3370–3378.
- Zhang Y, Yu ZF, Xu J, Zhang KQ. 2011. Divergence and dispersal of the nematode-trapping fungus Arthrobotrys oligospora from China. Environ Microbiol Rep. 3:763–773.
