Abstract
The complete mitochondrial genome (mitogenome) of Gryposmylus pennyi is described in the present paper. The sequenced mitogenome is 16,255 bp in length containing the typical set of 37 genes, including 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one control region. Besides, COI initiates with TCG, remaining 12 PCGs use ATN as the start codon. Most of PCGs terminate with TAA codons but COII, CytB, ND4 andND5 with a single T residue. The lrRNA and srRNA genes are 1331 bp and 793 bp in length, respectively. The control region is 1335 bp long with A + T content of 92.2%. All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGN). The phylogenetic result supports the monophyly of the family Osmylidae and a closer relationship between Gryposmylus and Heterosmylus that belong to the same subfamily Protosmylinae.
Gryposmylus Krüger, 1913 is a small genus within Protosmylinae (Neuroptera, Osmylidae) including only two species (Winterton and Wang Citation2016). The Oriental species Gryposmylus pennyi Winterton and Wang (Citation2016) shows the similar wing pattern to another Neotropical chrysopid species Vieira leschenaulti (Navás, 1911), which was regarded regarded as a dramatic convergence (Winterton and Wang Citation2016). In this study, the complete mitochondrial genome (mitogenome) of G. pennyi was reported and the phylogenetic relationship was reconstructed combining the current mitogenome data of other two osmylid species and other five families of Neuroptera.
The specimen was collected from Yinshan Park at Mountain Dayao in Guangxi province, China. Voucher specimen (No. CAU-Ogp-L2) was deposited at the Entomological Museum of China Agricultural University (CAU). The complete mitogenome was obtained using next-generation sequencing method with Illumina Hiseq 2500, and the sequence was deposited in GenBank under the accession number MK341582.
The whole mitogenome is a circular DNA molecule with 16,255 bp in length and contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and one control region. The gene arrangement is consistent with the putative ancestor of insects and also two other osmylid species, T. langii and Heterosmylus sp. (Boore Citation1999; Zhao et al. Citation2013; Wang et al. Citation2017). The nucleotide composition of the mitogenome is significantly biased towards A + T (80.5%) with negative AT-skew (−0.024) and negative GC-skew (−0.138). Twelve protein-coding genes initiate with ATN as the start codon (ATA for ND5; ATG for ND2, ATP6, COII, COIII, CytB, ND4, ND4L; ATT for ATP8, ND1, ND3, ND6;) except for the COI with TCG as the start codon. The typical termination codon (TAA) occurs in nine protein-coding genes and the remaining PCGs including COII, CytB, ND4 and ND5 terminate with a single T residue.
All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGN), which lacks a dihydroxyuridine (DHU) arm. The lrRNA is 1331 bp in length with an A + T content of 82.9% and the srRNA is 793 bp in length with an A + T content of 81.6%. The control region located between the srRNA and tRNAIle is 1335 bp with A + T content of 92.2%.
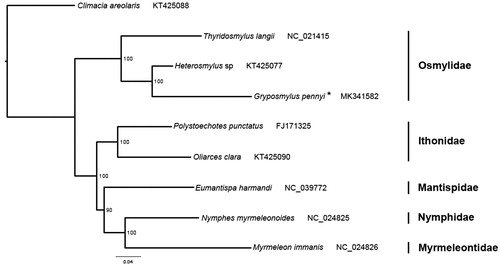
Phylogenetic tree was constructed based on the sequences of 13 protein-coding genes and two rRNA genes from nine neuropteran species using RAxML-HPC2 (Stamatakis Citation2014) with maximum-likelihood (ML) method under the GTR + CAT model () (Song et al. Citation2016; Li et al. Citation2017; Liu et al. Citation2018). The result supports the monophyly of the family Osmylidae and the grouping of Gryposmylus and Heterosmylus that belongs to the subfamily Protosmylinae. This phylogenetic relationship is in accordance with the current phylogeny of Osmylidae (Winterton et al. Citation2017).
Figure 1. Phylogenetic tree inferred from ML analysis of the nucleotide of the 13 PCGs and two rRNA genes (13162 bp). The nodal numbers indicate the bootstrap values obtained with 1000 replicates. Genbank accession numbers for the sequences are indicated next to the species names. The newly sequenced mitogenome is indicated by the asterisk.
Disclosure statement
All authors have read and approved the final manuscript. The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Li H, Leavengood JM Jr, Chapman EG, Burkhardt D, Song F, Jiang P, Liu JP, Zhou XG, Cai WZ. 2017. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of truebugs. Proc R Soc B Biol Sci. 284:20171223.
- Liu YQ, Song F, Jiang P, Wilson JJ, Cai WZ, Li H. 2018. Compositional heterogeneity in true bug mitochondrial phylogenomics. Mol Phylogenet Evol. 118:135–144.
- Song F, Li H, Jiang P, Zhou XG, Liu JP, Sun CH, Vogler A, Cai WZ. 2016. Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol Evol. 8:1411–1426.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang YY, Liu XY, Garzón‐Orduña IJ, Winterton SL, Yan Y, Aspöck U, Aspöck H, Yang D. 2017. Mitochondrial phylogenomics illuminates the evolutionary history of Neuropterida. Cladistics. 33:617–636.
- Winterton SL, Wang YJ. 2016. Revision of the genus Gryposmylus Krüger, 1913 (Neuroptera, Osmylidae) with a remarkable example of convergence in wing disruptive patterning. ZooKeys. 617:31–45.
- Winterton SL, Zhao J, Garzón-Orduña IJ, Wang YJ, Liu ZQ. 2017. The phylogeny of lance lacewings (Neuroptera: Osmylidae). Syst Entomol. 42:555–574.
- Zhao J, Li H, Winterton SL, Liu ZQ. 2013. Ancestral gene organization in the Mitochondrial genome of Thyridosmylus langii (McLachlan, 1870) (Neuroptera: Osmylidae) and implications for lacewing evolution. PLoS One. 8:e62943.
