Abstract
Here, we present complete mitochondrial genome of the four-lined snake, Elaphe quatuorlineata (Bonnaterre, 1790) and its comparison with closely related E. sauromates (Pallas, 1814). The complete sequence of the mitogenome is 17,183 bp long and consists of 13 protein-coding genes, two rRNA and 22 tRNA genes, and two control regions. It possesses the same gene order as other mitogenomes of Elaphe spp. and the base composition is as follows: 35.0% (A), 26.7% (C), 25.6% (T), and 12.7% (G), with an A + T bias (60.6%). The presented mitogenome will provide new data for phylogenetic analysis within phylogenetically close western Palearctic species of the genus Elaphe.
The four-lined snake, Elaphe quatuorlineata (Bonnaterre, 1790) is an endemic European species with a disjunct areal comprising the Apennine and Balkan peninsulas (Sindaco et al. Citation2013). This species comprises several morphologically distinct subspecies that more or less correspond to published molecular phylogeny based on mtDNA (Kornilios et al. Citation2014); these authors observed the highest genetic diversity in the Aegean region. On the other hand, populations from continental Balkans formed same phylogenetic clade as those from Italy. As its related species, E. sauromates (Pallas, 1814) was for a long time considered to be a subspecies of E. quatuorlineata; here, we provide their full mitogenome comparison.
Tissue sample (blood from adult female; no. 1509 stored in the collection of tissue samples at the Department of Zoology, Comenius University in Bratislava, Slovakia) was collected near Crkvino (41.66°N, 21.82°E), Republic of Northern Macedonia. Total genomic DNA was isolated by commercial DNA extraction kit (DNeasy® Blood and Tissue Kit, Qiagen) according to the manual. For data analysis with CLC Genomics Workbench 11.0 (https://www.qiagenbioinformatics.com) app., 3.5 million of paired-end reads were first trimmed of adapter sequences and low-quality bases. Remaining 2.8 million paired-end reads were mapped to the mitochondrial genome of the E. bimaculata (KM065513), resulting in the coverage of 97% of the reference sequence with 11.9x average coverage depth. The consensus sequence of E. quatuorlineata mtDNA was obtained from the mapped reads by the ‘‘Extract Consensus Sequence’’ tool executed with the following options: insert ‘‘N’’ for uncovered regions and simple voting for conflict resolution.
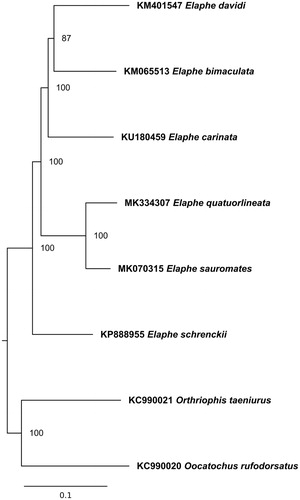
The phylogenetic analysis of the whole mitogenome sequences of all available Elaphe species was performed (). They were aligned using multiple sequence alignment program Muscle 3.8.3.1 (Edgar Citation2004). All gaps and poorly aligned positions were removed using Gblocks 0.91b (Talavera and Castresana Citation2007), resulting in 16,920 bp length alignment. The phylogenetic relationships were reconstructed using the maximum likelihood (ML) method in the PhyML 2.4.5 (Guindon and Gascuel Citation2003) and tested by 1000 bootstrap replications. The best substitution model was selected by the jModelTest 2.1.10 (Darriba et al. Citation2012) based on the Bayesian information criterion (BIC).
Figure 1. Maximum likelihood phylogenetic tree of Elaphe representatives. The tree was created using TIM2 + G model. Mitogenomes of Orthriophis taeniurus (Cope, 1861) and Oocatochus rufodorsatus (Cantor, 1842) were used as an outgroup. GenBank accession numbers and bootstrap values of nodes are shown on the tree.
The complete mitochondrial genome of E. quatuorlineata is 17,183 bp long (GenBank accession number: MK334307) and contains 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and two control regions. The overall base composition of the mitogenome in descending order was 35.0%—A, 26.7%—C, 25.6%—T, 12.7%—G, with an A + T bias (60.6%), that was almost identical to E. sauromates (34.8%—A, 26.8%—C, 25.7%—T, 12.7%—G).
Ten of the 13 PCGs (ATP8, ATP6, COX2, COX3, CYTB, ND1, ND3, ND4, ND4L, ND5,) used ATG as start codon, while COX1 used GTG, ND2 used ATT, and ND6 used ATA. The only difference in start codon usage compared to E. sauromates is for ND6 (ATG in E. sauromates). Eleven genes (ATP8, ATP6, COX2, COX3, CYTB, ND1, ND2, ND3, ND4L, ND4, and ND5) ended with a TAA stop codon, but for six of them (COX2, COX3, CYTB, ND1, ND2, and ND3) TAA stop codon is completed by the addition of 3' A residues to the mRNA in both species. Here, ND6 gene ended with an AGA and COX1 with AGG stop codon, whereas in E. sauromates these genes have different stop codons (ND6—AGG, COX1—AGA).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Kornilios P, Thanou E, Lymberakis P, Sindaco R, Liuzzi C, Giokas S. 2014. Mitochondrial phylogeography, intraspecific diversity and phenotypic convergence in the four-lined snake (Reptilia, Squamata). Zoolog Scripta. 43:149–160.
- Sindaco R, Venchi A, Grieco C. 2013. The reptiles of the western Palearctic 2. Annotated checklist and distributional Atlas of the Snakes of Europe, North Africa, Middle East and Central Asia. Latina (Italy): Edizioni Belvedere.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577.
