Abstract
The complete mitochondrial genomes of the Spotted Anole (Anolis punctatus), Florida Scrub Lizard (Sceloporus woodi), and Mesquite Lizard (S. grammicus) were assembled and annotated. Genomic DNA of A. punctatus was sequenced in one-sixth of a lane in an Illumina HiSeq while comparable sequencing data for S. woodi and S. grammicus were retrieved from the Short Read Archive (entries SRR1286259 and SRR1145766, respectively). We assembled these mitogenomes using MIRA, NovoPlasty, and/or MITObim software. The resulting whole and circularized mitogenome assemblies exhibited typical vertebrate sequence length and gene organization. We deposited these mitogenomes into public databases under accession numbers MK091854, BK010487, and BK010486, respectively. To verify the taxonomic identifications of our mitogenomes (as iguanian lizards), we conducted maximum likelihood analyses with 14 published mitogenomes from the lizard superfamily Iguanoidea and two outgroups. Our analysis of concatenated gene sequences from mitochondria confirmed the taxonomic identities of our mitogenomes to the genus level.
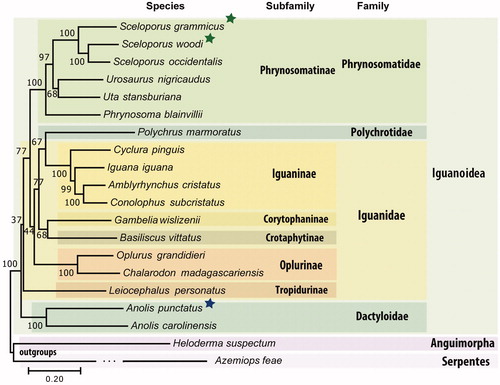
With more than 400 species described, the genus Anolis represents one of the most speciose vertebrate genera (Losos Citation2009). However, to date, only one complete mitogenome is available for Anolis. The Spotted Anole, Anolis punctatus is widespread in Amazonia and coastal Atlantic Forests of South America (Ribeiro-Júnior Citation2015; Prates et al. Citation2016). The sample sequenced here (MNRJ 23455) was obtained from the reptile collection at the Museu Nacional, Universidade Federal do Rio de Janeiro. The partial genome of A. punctatus was sequenced in one-sixth of a lane in an Illumina HiSeq and produced ∼50 million 100-bp paired-end sequencing reads. Spiny lizards from the genus Sceloporus are represented by ∼90 species in North America (Leaché Citation2010). Like Anoles, Sceloporus is only represented by one mitogenome in Genbank. We assembled the mitogenome of the Florida Scrub Lizard (Sceloporus woodi) endemic to Florida (Demarco Citation1992) and the Mesquite Lizard Sceloporus grammicus from Northern and Central Mexico (Arévalo et al. Citation1991; Ochoa-Ochoa and Villella 2006). Illumina datasets were downloaded from the Short Read Archive (SRR1286259 for S. woodi and SRR1145766 for S. grammicus) originally obtained from specimens UWBM 7265 and UWBM 6585 in the Burke Museum of Natural History and Culture, University of Washington (Leaché et al. Citation2013; Genomic Resources Development Consortium Citation2015). For the Spotted Anole, a de novo assembly using MIRA software (Chevreux et al. Citation1999) produced a nearly complete version for the mitogenome. An additional assembly step was needed to confirm circularization. The complete A. punctatus mitochondrial genome is 17,132 bp and was produced using 18,623 sequencing reads with an average read coverage of 91× according to Tablet software (Milne et al. Citation2013). For S. woodi, the mitogenome was produced using a reference-based MIRA job guided by the mitochondrial genome of S. occidentalis (NC_005960.1) before being completed by MITObim (Hahn et al. Citation2013). For S. grammicus, the raw sequence reads were used as input for Novoplasty 2.6.3 (Dierckxsens et al. Citation2016) and finalized using MITObim. The complete and circularized mitogenome for S. woodi is 17,301 bp based on 26,225 reads while the S. Grammicus mitogenome is 16,830 bp based on 5832 reads. Tablet indicated that the average coverages of S. woodi and S. grammicus mitogenomes were ∼483× and ∼203×. Notably, we found several high-coverage regions in our mitogenome assemblies, which may indicate the presence of mitochondrial pseudogenes (NuMTs). These high-coverage regions are: 13,650-13,914 and 15,173-15,542 in A. punctatus; 15,419-15,568 in S. grammicus; and 5,114-7,020 and 11,611-13,216 in S. woodi. Automatic annotation of mitogenomes was performed using the MITOS Web Server (Bernt et al. Citation2013) and geneCheker (Schomaker-Bastos and Prosdocimi, Citation2018), followed by manual curation. The annotated mitogenomes were submitted to GenBank and TPA databases (A. punctatus: MK091854; S. woodi: BK010487; S. grammicus: BK010486). To confirm the taxonomic identities, we performed a phylogenetic analysis using all 14 complete mitogenomes available for the Iguanoidea superfamily and two outgroups. Phylomito software (https://github.com/igorrcosta/phylomito) extracted the protein-coding genes, performed sequence alignments, and concatenations. A maximum likelihood analysis confirmed monophyly for Anolis and Sceloporus genera ().
Figure 1. Phylogenomics tree inferred using maximum likelihood based on the General Time Reversible model in MEGA7. Genes from complete mitochondrial genomes from all available species from the Iguanoidea superfamily were downloaded, aligned, and concatenated into a dataset composed of 11,505 nucleotide positions. Bootstrap values (1000 replicates) are shown at the corresponding nodes. Colored boxes indicate clades described, green asterisks represent new mitochondrial genomes described here using public data while blue asterisk represent the new mitochondrial genome described here using our own data. The accession numbers of the mitogenomes used in the tree are BK010486.1, BK010487.1, NC_005960.1, NC_026308.1, NC_027261.1, NC_036492.1, NC_012839.1, NC_027089.1, NC_002793.1, NC_028031.1, NC_028030.1, NC_012831.1, NC_012829.1, NC_012827.1, NC_012836.1, NC_012834.1, MK091854.1, NC_010972.2, NC_008776.1, and NC_030781.1.
Acknowledgements
We would like to thank Adam Leaché for providing helpful information about the origins of the Sceloporus sequences used in this study. We cherish the inspiring presence of Alex Schomaker (in memoriam) that worked in initial versions of the anole mitogenome: thank you, Alex.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Arévalo E, Porter CA, González A, Mendoza F, Camarillo JL, Sites JW, Arevalo E, Gonzalez A, Camarillo JL. 1991. Population cytogenetics and evolution of the Sceloporus grammicus complex (Iguanidae) in central Mexico. Herpetol Monogr. 5:79–115.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc German Conf Bioinform. 99:45–56.
- Demarco V. 1992. Florida scrub lizard. In Moler PE, editor. Rare and endangered biota of Florida, Vol. III, amphibians and reptiles. Gainesville (FL): University of Florida Press; p. 141–145.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Genomic Resources Development Consortium. 2015. Genomic resources notes accepted 1 August 2014–30 September 2014. Mol Ecol Res. 15:228–229.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucl Acids Res. 41:e129.
- Leaché AD. 2010. Species trees for spiny lizards (genus Sceloporus): identifying points of concordance and conflict between nuclear and mitochondrial data. Mol Phylogenet Evol. 54:162–171.
- Leaché AD, Harris RB, Maliska ME, Linkem CW. 2013. Comparative species divergence across eight triplets of spiny lizards (Sceloporus) using genomic sequence data. Genome Biol Evol. 5:2410–2419.
- Losos JB. 2009. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley: University of California Press.
- Milne I, Stephen G, Bayer M, Cock PJA, Pritchard L, Cardle L, Shaw PD, Marshall D. 2013. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 14:193–202.
- Ochoa-Ochoa LM, Villela OF. 2006. Áreas de diversidad y endemismo de la herpetofauna mexicana. Distrito Federal (México): Universidad Nacional Autónoma de México and Comisión Nacional para el Conocimiento y Uso de la Biodiversidad.
- Prates I, Xue AT, Brown JL, Alvarado-Serrano DF, Rodrigues MT, Hickerson MJ, Carnaval AC. 2016. Inferring responses to climate dynamics from historical demography in neotropical forest lizards. Proc Natl Acad Sci USA. 113:7978–7985.
- Ribeiro-Júnior, Marco A. 2015. Catalogue of distribution of lizards (Reptilia: Squamata) from the Brazilian Amazonia. I. Dactyloidae, Hoplocercidae, Iguanidae, Leiosauridae, Polychrotidae, Tropiduridae. Zootaxa 3983:1–110.
- Schomaker-Bastos A. Prosdocimi F. mitoMaker: A Pipeline for Automatic Assembly and Annotation of Animal Mitochondria Using Raw NGS Data. Preprints 2018, 2018.08.0423. doi:10.20944/preprints201808.0423.v1.
