Abstract
The complete mitogenome of Gryllus bimaculatus is 16,301 bp in length, contains the typical set of 37 genes, including 13 protein-coding genes (PCGs) (ATP6, ATP8, COI-III, ND1-6, ND4L, and Cyt b), two rRNAs (12S rRNA and 16S rRNA), 22 tRNAs, and a putative CR (D-loop). ATT, ATG, and ATA are the most start codon. TAA is the most frequent stop codon. The new mtDNA sequence contains 16S rRNA and 12S rRNA of rRNA, which is located between tRNALeu and D-loop, separated by tRNAVal. All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGN) and tRNALeu(CUN), which lacks a dihydroxyuridine (DHU) arm. The D-loop (1585 bp) located the end of mitochondrial genome sequences. The phylogenetic trees support Chiromantes haematochir has close relative with Velarifictorus hemelytrus and Teleogryllus emma.
Gryllus bimaculatus is known as the common name two-spotted cricket, it can be discriminated from other Gryllus species by the two dot-like marks on the base of its wings (Miyashita et al. Citation2016). The species is one of field cricketsis, belong to family Gryllidae (superfamily Grylloidea), which is as pet in China. The sample of G. bimaculatus was collected from Zhanjiang City (110°55'08.10'' E, 21°27'07.02'' N), Guangdong Province in May, 2018. The sample was stored in the Biology Specimen Room of the College of Life Sciences, Anhui Medical University, China (Sample code is AMHU-XS20181102). The DNA sample was stored in the −80 °C in the Cancer Cell Biology Laboratory, College of Life Sciences, Anhui Medical University. The complete mtDNA sequence of G. bimaculatus has been assigned GenBank accession number MK204367.
The length of the complete mtDNA sequence is 16,301 bp, similar to other Gryllidae species (Cha et al. Citation2007; Fenn et al. Citation2007; Yang et al. Citation2016). It contains the typical set of 37 gene, including 13 protein-coding genes (ND2, COI-III, ATP8, ATP6, ND1-6, ND4L, and Cyt b), two rRNAs (12S rRNA and 16S rRNA), 22 tRNAs and a putative D-loop. The overall base composition for the mtDNA sequence is as follows: A, 40.5%, C, 17.0%, G, 9.7%, T, 32.8%. The new sequence has higher A + T content (73.3%) and lower C + G content (26.7%), similar to other invertebrate species (Cha et al. Citation2007; Fenn et al. Citation2007; Yang et al. Citation2016). Through the 13 PCGs, the longest one is ND5 (1721 bp), and the shortest is ATP8 (162 bp), similar with the other species (Cha et al. Citation2007; Fenn et al. Citation2007; Yang et al. Citation2016). ATT, ATG, and ATA are the most start codon in all PCGs except COI start with GTA and ND3 start with TCG. TAA is the most frequent stop codon although COI and ND6 end with TA-, COII and ATP6 stop with the single nucleotide T-, and ND3, ND4, and ND5 stop with the single nucleotide TAG. The new mtDNA sequence contains 16S rRNA and 12S rRNA of rRNA, which is located between tRNALeu and D-loop, separated by tRNAVal. The 12S rRNA is 718 bp long and the 16S rRNA is 1333 bp in length. All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGN) and tRNALeu(CUN), which lacks a dihydroxyuridine (DHU) arm. The non-coding regions include a control region (D-loop) and a few intergenic spacers. The D-loop, which is located the end of mitochondrial genome sequences, is 1585 bp long.
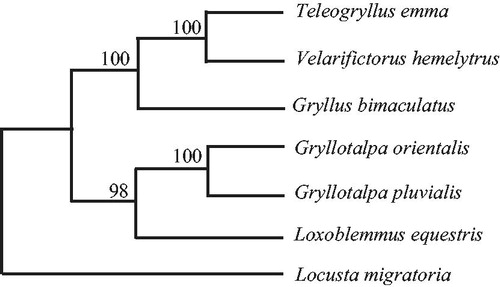
Phylogenetic trees were estimated using ML and BI methods, based on the complete mtDNA of six crickets, and corresponding Locusta migratoria (NC_001712) sequence was used as outgroup, sharing similar topologies and high node support values (). Chiromantes haematochir has close relative with Velarifictorus hemelytrus and Teleogryllus emma.
Figure 1. Phylogenetic relationships among the six crickets based on complete mtDNA sequences. Numbers at each node are maximum likelihood bootstrap proportions (estimated from 100 pseudoreplicates). The accession number in GenBank of six crickets in this study: Loxoblemmus equestris (KU562919), Gryllotalpa pluvialis (EU938371), Gryllotalpa orientalis (AY660929), Gryllus bimaculatus (MK204367), Velarifictorus hemelytrus (KU562918), and Teleogryllus emma (NC_001712).
Nucleotide sequence accession number
The complete mtDNA sequence of G. bimaculatus has been assigned with GenBank accession number MK204367.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. 2007. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 392:206–220.
- Fenn JD, Cameron SL, Whiting MF. 2007. The complete mitochondrial genome sequence of the Mormon cricket (Anabrus simplex: Tettigoniidae: Orthoptera) and an analysis of control region variability. Insect Mol Biol. 16:239–252.
- Miyashita A, Kizaki H, Sekimizu K, Kaito C. 2016. No effect of body size on the frequency of calling and courtship song in the two-spotted cricket, Gryllus bimaculatus. PLoS One. 11:e0146999.
- Yang J, Ren Q, Huang Y. 2016. Complete mitochondrial genomes of three crickets (Orthoptera: Gryllidae) and comparative analyses within Ensifera mitogenomes. Zootaxa. 4092:529–547.
