Abstract
The complete mitochondrial genome sequence of the hybrid fish Salvelinus fontinalis (♀) × Salvelinus malma sp. (♂) was sequenced. It was 16,623-nucleotide in length and consisted of 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 2 non-coding regions (L-strand replication origin and control region), showing conserved gene arrangement with most vertebrates. The phylogenetic tree showed the hybrid salmon to be one of the Salvelinus and the relationships of S. fontinalis were closer. The complete mitochondrial genome sequence of S. fontinalis (♀) × S. malma sp. (♂) provided an important dataset for a better understanding of the mitogenomic diversities and evolution in salmon fish as well as novel genetic markers for studying population genetics and species identification.
In animal evolution, hybridization is a useful means of inducing genetic and epigenetic alterations leading to increased diversity and speciation (Barton Citation2001). Animal mitochondrial DNA (mtDNA) is almost exclusively inherited from the mother (White et al. Citation2008); however, Luo et al. said they found mtDNA parental inheritance in three families (Luo et al. Citation2018). There is no report of the complete genome of this hybrid fish Salvelinus fontinalis (♀) × Salvelinus malma sp. (♂), which was developed in Tonghua, Jilin Province, Republic of China (N41°35′29″, E127°26′34″, altitude 680 m, air pressure 93.59 kpa) (Doiron et al. Citation2002; Yang et al. Citation2017). Therefore, it is very important to characterize the complete mitogenome of this species, which can be utilized in research on taxonomic resolution, population genetic structure and phylogeography, and phylogenetic relationship. Total DNA was extracted from muscle following TIANamp Marine Animals DNA Kit (Tiangen, China). Totally 24 pairs of primers were used, in which 16 pairs of primers were retrieved from Gonostoma gracile (Miya and Nishida Citation1999) and the remaining primers were designed based on public mitochondrial genome sequences of S. fontinalis. The samples were stored in −80 °C in Research Institute of Marine Biotechnology and life Health, Ningbo University, Ningbo.
In this study, we obtained the complete mitochondrial genome of the hybridized fish S. fontinalis (♀) × S. malma sp. (♂). Its mitochondrial genome has been deposited in the GenBank under accession number MK270623. For a better understanding of the genetic status and the evolutionary study, we focused on the genetic information contained in the complete mitochondrial genomes of the fish.
The complete mitogenome of the hybrid fish S. fontinalis (♀) × S. malma sp. (♂) was 16,623 bp in length. The genomic organization was identical to those of typical vertebrate mitochondrial genomes, including 2 rRNA genes, 13 protein-coding genes, 22 tRNA genes, a light-strand replication origin (OL), and a putative control region (CR). The overall base composition was 28.3% of A, 26.5% of T, 28.4% of C, and 16.8% of G with a slight A + T bias (54.8%) like other vertebrate mitochondrial genomes. The features mentioned above were accordant with typical Salvelinus fish mitogenome.
For the 13 protein-coding genes, 12 genes started with ATG while only COI started with GTG. Five genes shared the termination codon TAA (COI, ATPase8, ND1, ND5, and ND4L), one with TAG (ND6), the remaining with incomplete stop codon (COII, COIII, ND2, ND3, ND4, ATPase6, and Cytb). This feature was common among vertebrate mitochondrial protein-coding genes. Salvelinus fontinalis (♀) × S. malma sp. (♂) had two non-coding regions, the L-strand replication origin region (36 bp) was located between tRNA-Asn and tRNA-Cys, and the control region (962 bp) located within the tRNA-Pro and tRNA-Phe. Eight tRNA (tRNA-Ser, tRNA-Pro, tRNA-Glu, tRNA-Tyr, tRNA-Cys, tRNA-Asn, tRNA-Ala, and tRNA-Gln) and the ND6 gene were encoded on the L-strand, the others were encoded on the H-strand. This feature is similar to other fish mitochondrial genes. The complete mitogenome sequence had 16s RNA (1678 bp) and 12s RNA (946 bp), which were located between tRNA-Phe and tRNA-Leu and separated by tRNA-Val gene. The location is same with most vertebrates that have high conservation.
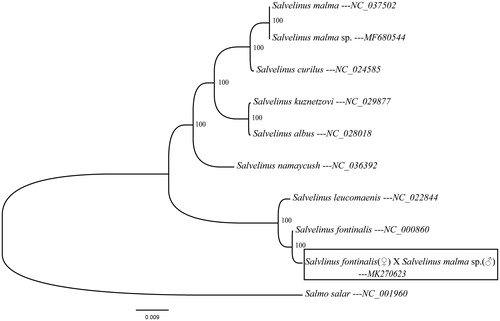
To determine the taxonomic status of the hybrid salmon, we reconstructed the phylogeny of this salmon stock with other natural populations in salmon based on the entire mitogenome sequences. The phylogenetic tree showed the hybrid salmon to be one of the Salvelinus and the closer relationship with S. fontinalis other than S. malma sp. (). The phylogeny was reconstructed based on the General Time Reversible + gamma sites (GTR + G) model of nucleotide substitution using Mega7 (Kumar et al. Citation2016). The complete mitochondrial genome sequence of the S. fontinalis (♀) × S. malma sp. (♂) hybrid provided an important dataset for a better understanding of the mitogenomic diversities and evolution in salmon fish as well as novel genetic markers for studying population genetics and species identification.
Figure 1. Maximum likelihood tree for the S. fontinalis (♀) S. malma sp. (♂), and the GenBank representatives of the family Salmonidae. The tree is constructed using whole mitogenome sequences. The tree is based on the general time reversible + gamma sites (GTR + G) model of nucleotide substitution. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barton NH. 2001. The role of hybridization in evolution. Mol Ecol. 10:551–568.
- Doiron S, Bernatchez L, Blier PU. 2002. A comparative mitogenomic analysis of the potential adaptive value of Arctic charr mtDNA introgression in brook charr populations (Salvelinus fontinalis Mitchill). Mol Biol Evol. 19:1902–1909.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Luo S, Valencia CA, Zhang J, Lee NC, Slone J, Gui B, Wang X, Li Z, Dell S, Brown J, et al. 2018. Biparental inheritance of mitochondrial DNA in humans. Proc Natl Acad Sci USA. 115:13039–13044.
- Miya M, Nishida M. 1999. Organization of the mitochondrial genome of a deep-sea fish, Gonostoma gracile (Teleostei:Stomiiformes): first example of transfer RNA gene rearrangements in bony fishes. Mar Biotechnol. 1:416–426.
- White DJ, Wolff JN, Pierson M, Gemmell NJ. 2008. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 17:4925–4942.
- Yang L, Meng F, Wang R, Shi G. 2017. Complete mitochondrial genome of the Salvelinus malma sp. (Salmoniformes, Salmonidae) with phylogenetic consideration. Mitochondrial DNA Part B. 2:889–890.
