Abstract
Endemic to the southeast boundary of Tibet plateau, the medicinal alpine shrub Caragana kozlowii is a threatened species. The genetic and molecular data about it is insufficient. In this paper, the complete chloroplast (cp) genome sequence of C. kozlowii was sequenced by the Illumina sequencing platform. The result suggests that C. kozlowii cp genome is relatively short because it lacks an Inverted Repeat (IR) region. It was 131,274 bp in length, containing one large and one small single copy region (SSC and LSC) of 85,281 and 18,919 bp separated by an inverted repeat region (IR) of 27,074 bp, respectively. It contains unique 110 genes, including 76 protein-coding genes, 30 tRNAs, and 4 rRNAs. Similar to the other Inverted Repeat-Lacking Clade (IRLC) plastomes, rpl22 and rps16 are absent. The cp genome has 34.5% GC content. Phylogenetic analysis based on ten chloroplast genomes indicates that C. kozlowii is closely related to C. rosea.
Caragana kozlowii Kom., which belongs to the family Fabaceae, is narrowly distributed in the southeast boundary of Tibet plateau (Chang et al. Citation2007). This species is an important medicinal plant, with its roots and flowers used in traditional Chinese medicine for their antineuralgic and antiarthritic properties (Meng et al. Citation2009). Because of the harsh environment and over-exploitation, the wild resources of C. kozlowii have dramatically decreased and only two sampling localities were found in the east of Tibet Autonomous Region. It is of utmost importance to protect, and genetically study the threatened species. However, there are few DNA sequences available for C. kozlowii in the GenBank. In this paper, we determined and analyzed the chloroplast (cp) genome of C. kozlowii, which will provide comprehensive information relevant to the effective conservation and management of this medicinal alpine shrub.
Fresh leaves were collected from C. kozlowii in Chengdu (31°17’N, 97°22’E), Tibet, China. Genomic DNA was extracted with a DNeasy Plant Kit (QIAGEN, Valencia, California, USA). The voucher specimen and DNA sample of C. kozlowii were deposited in the Molecular Ecology Laboratory (MEL), Institute of Loess Plateau, Shanxi University (Taiyuan, Shanxi, China). Then, Genomic DNA was sequenced on the Illumina Hiseq Platform (Illumina, San Diego, CA). ∼3.1 GB of raw data were generated with 150 bp pair-end read length. we then filtered and assembled the complete cp genome using the program NOVOPlasty (Dierckxsens et al. Citation2016), The resulting contigs were linked based on overlapping regions after being aligned to Caragana microphylla (KX289922) (Liu et al. Citation2016) and visualized in Geneious R10 (Biomatters Ltd., Auckland, New Zealand). The complete cp genome was annotated with the program Plann (Huang and Cronk Citation2015) and submitted to GenBank with the accession number MG386382. A physical map of the cp genome was generated by OGDRAW (http://ogdraw.mpimp-golm.mpg.de/).
The cp genome is relatively short because it lacks an Inverted Repeat (IR) region. It was 131,274 bp in length, containing one large and one small single copy region (SSC and LSC) of 85,281 and 18,919 bp separated by an inverted repeat region (IR) of 27,074 bp, respectively. It contains unique 110 genes, including 76 protein-coding genes, 30 tRNAs and 4 rRNAs. Similar to other IRLC plastomes, rpl22 and rps16 are absent. The cp genome has 34.5% GC content.
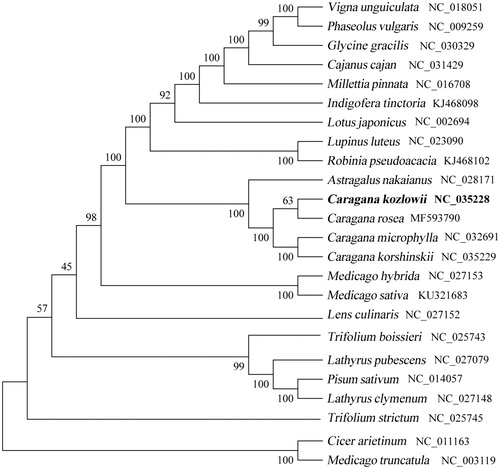
To confirm the phylogenetic position of C. kozlowii, the complete cp genome and 23 reported plastome were aligned by MAFFT (Katoh and Standley Citation2013) and a Neighbor-Joining (NJ) phylogenetic tree was constructed in RaxML version 8 (Stamatakis Citation2014). The phylogenetic tree showed that C. kozlowii is closely related to the congeneric C. rosea ().
This complete cp genome can be selected to develop highly variable molecular markers for population genetic survey, genetic relationships characterization, and other systematic studies. Hence, our study will be benefited for formulating potential new conservation and management strategies for this endangered medicinal alpine shrub.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chang ZY, Xu LR, Shi FC. 2007. A taxonomic revision of Caragana tangutica and C. kozlowi (Leguminosae). Acta Phytotaxonomica Sinica. 45:391–395.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl in Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol and Evol. 30:772–780.
- Liu B-b, Duan N, Zhang H-l, Liu S, Shi J-W, Chai B-F. 2016. Characterization of the whole chloroplast genome of Caragana microphylla Lam (Fabaceae). Conserv Genet Res. 8:371–373.
- Meng Q, Niu Y, Niu X, et al. 2009. Ethnobotany, phytochemistry and pharmacology of the genus Caragana used in traditional Chinese medicine. J Ethnopharmacol. 24:350–368.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.