Abstract
The complete sequence of the mitochondrial DNA of Nemoura papilla, has been completed and annotated in this study. The circular genome is 15,774 bp in length with an A + T content of 68.0% and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNAs), and a putative control region. All tRNA genes have the typical cloverleaf structure, with an exception for tRNASer(AGN). All PCGs use normal start codon ATN, while ND1 use TTG as the start codon. Meanwhile, 11 PCGs use the typical termination codons TAN, except COII and ND5, which stopped with the incomplete termination signal T. Phylogenetic analysis suggests that N. papilla is closely related to N. nankinensis. Nemouridae + Capniidae was assigned to be the sister of Taeniopterygidae.
Nemouridae is one of the largest families of Plecoptera with approximately 400 species (Baumann Citation1975; DeWalt et al. Citation2018). The stonefly (Nemoura papilla) inhabits transboundary streams and rivers of northern China, south of the Russian Far East, Japan, and Korea (Teslenko Citation2016; DeWalt et al. Citation2018). Though the phylogenetic position of Nemouridae in the superfamily Nemouroidea has been conducted based on morphological data (Zwick Citation2000), several different suggestions were proposed based on molecular data (Chen and Du 2018; Wang et al. Citation2019). In this study, we assembled the mitochondrial genome of N. papilla (GenBank accession no. MK290826) by high-throughput sequencing method for the first time.
The male adult sample of N. papilla was collected from Long Yu Bay Scenic Area, Henan Province, China in 2016. The thorax muscle of the specimen was used to extract total genomic DNA. Voucher specimens were deposited in Henan Institute of Science and Technology, China. The mitogenomes were amplified and sequenced as described in previous studies (Li et al. Citation2017; Wang et al. Citation2017; Liu et al. Citation2018). Sequence reads were assembled into contigs with BioEdit version 7.0.5.3 (Hall Citation1999). tRNA genes were identified by ARWEN (Laslett and Canbäck Citation2008) with the default setting. Two rRNA and all PCGs genes were identified alignment with homologous genes from other published stonefly mitogenomes.
The complete mitogenome of N. papilla is 15,774 bp in length with an A + T content of 68.0%. It encoded 37 genes including 13 PCGs, 22 tRNA genes, 2 rRNA genes, and a control region similar to most other insects. The gene order and organization of N. papilla are consistent with those of putative ancestor of insects (Boore Citation1999). The A + T content of PCGs, tRNAs, rRNAs, and control region was 65.7%, 70.3%, 72.2%, and 82.4%, respectively. Alignment of the sequences of the 13 PCGs was inferred from the amino acid alignment using ClustalW in MEGA 5.0 (Tamura et al. Citation2011). Most PCGs contained ATN codons. One exceptional was the ND1, which used TTG as a start codon, as reported for most other Plecoptera (Wang et al. Citation2017). Eleven PCGs used the typical termination codons TAA and TAG, while COII and ND5 stop with the incomplete terminaton signal T. All tRNAs could be folded into clover-leaf structures except the tRNASer(AGN). Both rRNAs were located on the N-strand of which was 1326 bp (lrRNA) long and 794 bp (srRNA) long, respectively. In addition to the control region, there were 73 nucleotides dispersed in 11 intergenic spacers. Gene overlaps were also found at fifteen gene junctions involving 50 nucleotides.
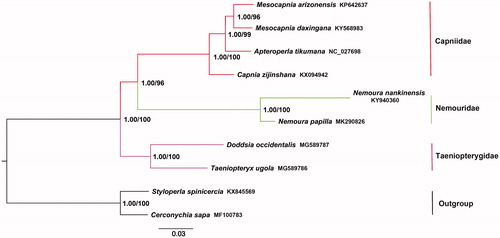
The phylogenetic trees among three families in the superfamily Nemouroidea were constructed from the maximum likelihood (ML) and Bayesian (BI) inferences based on the concatenated nucleotide sequences of PCG12 (contains the first and second codon of PCGs) dataset. The two methods exhibited the same tree topologies (). Phylogenetic analysis suggests that N. papilla is closely related to N. nankinensis. Nemouridae + Capniidae was assigned to be the sister of Taeniopterygidae. Our results support the traditional morphology-based cladistics (Zwick Citation2000).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baumann RW. 1975. Revision of the stonefly family Nemouridae (Plecoptera): a study of the world fauna at the generic level. Smithsonian Contributions to Zool. 40:1–84.
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Chen ZT, Du YZ. 2018. The first two mitochondrial genomes from Taeniopterygidae (Insecta: Plecoptera): structural features and phylogenetic implications. Int J Biol Macromol. 111:70–76.
- DeWalt RE, Maehr MD, Neu-Becker U, Stueber G. 2018. Plecoptera species file online. http://Plecoptera.SpeciesFile.org. [accessed 10 Dec 2018].
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Li H, Leavengood JJM, Chapman EG, Burkhardt D, Song F, Jiang P, Liu J, Zhou XG, Cai WZ. 2017. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc R Soc B. 284:1862.
- Liu YQ, Song F, Jiang P, Wilson JJ, Cai WZ, Li H. 2018. Compositional heterogeneity in true bug mitochondrial phylogenomics. Mol Phylogenet Evol. 118:135–144.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28:2731–2739.
- Teslenko VA. 2016. Larvae of two East-Asian species of Nemoura (Plecoptera: Nemouridae). Zootaxa. 4162:381–390.
- Wang Y, Cao JJ, Li WH. 2017. The complete mitochondrial genome of the styloperlid stonefly species Styloperla spinicercia Wu (Insecta: Plecoptera) with family-level phylogenetic analyses of the Pteronarcyoidea. Zootaxa. 4243:125–138.
- Wang Y, Cao JJ, Li N, Ma GY, Li WH. 2019. The first mitochondrial genome from Scopuridae (Insecta: Plecoptera) reveals structural features and phylogenetic implications. Int J Biol Macromol. 122:893–902.
- Zwick P. 2000. Phylogenetic system and zoogeography of the Plecoptera. Annu Rev Entomol. 45:709–746.