Abstract
The complete mitochondrial genome of Sinularia ceramensis was completed using next-generation sequencing (NGS) method. The mitochondrial genome is a circular molecule of 18,740 bp in length. The gene arrangements including 14 protein-coding genes (PCGs), 2 rRNAs, and 1 tRNA (tRNA-Met). The base composition is 30.26% A, 16.44% C, 16.35% G, and 33.95% T, with a G + C content of 35.76%. According to the phylogenetic analysis, Alcyonacea family are clustered in different clades.
The genus Sinularia May, 1898 is one of the most pervasive Indo-Pacific soft corals that has been settled in wide range of habitats (Fabricius and Alderslade Citation2001). So far, the complete mitogenome of two species of Sinularia consisting of Sinularia peculiaris (JX023274 and NC_018379) and Sinularia cf. cruciate (KY462727 and NC_034318) have already been performed. In the present study, the complete mitochondrial genome of Sinularia ceramensis Verseveldt, 1977 (GenBank: MK292119) was sequenced using next-generation sequencing.
An individual of S. ceramensis was collected from the South China Sea (Sanya, Hainan province, China; 18° 14′ 5.93″ N, 109° 22′ 46.46″ E) and stored in Hainan Tropical Ocean University Museum of Zoology. The specimen was identified using PuCAs- mtMutS (Benayahu et al. Citation2018). The genomic DNA was extracted by Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, CN; NO. B518221). A genomic library was constructed followed by paired-end (2 × 150 bp) next-generation sequencing, using the Illumina HiSeq X-ten sequencing platform (Tianjin, China). Quality checks for sequencing reads were carried out by FastQC (Andrews Citation2010) and the sequences were assembled and mapped to the reference Sinularia mitochondrial genome (Sinularia peculiaris, JX023274) with Spades v3.9.0 (Bankevich et al. Citation2012) and bowtie v2.2.9 (Langmead and Salzberg Citation2012). Putative tRNA gene was determined using ARWEN (http://130.235.46.10/ARWEN/) online software. All genes were annotated based on gene order on the reference mitochondrial map using BLAST analysis (https://blast.ncbi.nlm.nih.gov). In addition, to annotate PCGs, the position of start and stop codons were re-considered.
The complete mitogenome of S. ceramensis was 18,740 bp in length with 14 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), and 1 transfer RNA (tRNA-Met). The overall nucleotide composition of the major strand of the S. ceramensis mitogenome was as follows: 30.26% A, 16.44% C, 16.35% G, and 33.95% T, with a total G + C content of 35.76%.
The tRNA-Met and four protein-coding genes (COX3, ATP6, ATP8, and COX2) were located on the light strand. All PCGs began with common ATG start codon. Stop codons included TAG (ND1, ND6, ND3, ND2, ND5, COX3, and COX2), TAA (CYTB, ND4L, mutS, ND4, ATP6, and ATP8) and non-complete codons T- (COX1).
The 12S ribosomal RNA and 16S ribosomal RNA were encoded on the heavy strand from 1583 to 2633 (1051 bp) and 9157 to 11124 (1968 bp), respectively, with 12S having a rather lower A + T content (56.61% vs. 58.49%). The longest gap but a single overlap were determined between COX2/COX1 (112 bp) and ND2/ND5 (−13 bp), respectively.
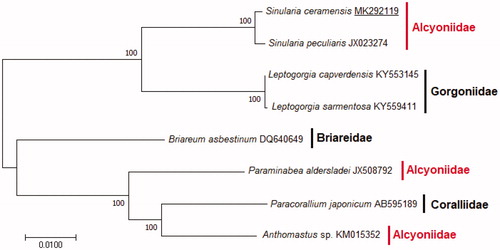
The phylogenetic relationship of S. ceramensis among order Alcyonacea was performed from a concatenated dataset consisting of the 14 PCGs and 2 rRNAs using the software MEGA 7.0.26 v. (Kumar et al. Citation2016) with 1000 bootstrap replicates and GTR model (). According to the result, Alcyoniidae was divided into three separated clades while families Gorgoniidae, Briareidae, and Coralliidae were placed between Alcyoniidae clades. A comprehensive taxonomical status of Alcyoniidae family needs to revise using whole mitochondrial genome sequences.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. (04-10-18: Version 0.11.8 released) http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Benayahu Y, van Ofwegen LP, Dai C-f, Jeng MS, Soong K, Shlagman A, Du SW, Hong P, Imam NH, Chung A, et al. 2018. The octocorals of Dongsha Atoll (South China Sea): an iterative approach to species identification using classical taxonomy and molecular barcodes. Zool Stud. 57:50.
- Fabricius K, Alderslade P. 2001. Soft corals and sea fans: a comprehensive guide to the tropical shallow-water genera of the Central-West Pacific, the Indian Ocean and the Red Sea. Townsville: Australian Institute of Marine Science.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33:1870–1874.
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods. 9:357–359.