Abstract
The complete chloroplast genome sequence of Artocarpus nanchuanensis was determined, an Endangered species of Artocarpus in the family Moraceae. The plastome is 160,752 bp in length, exhibiting a characteristic quadripartite structure with a pair of inverted repeat regions (IRs) of 25,693 bp, separated by a large single-copy region (LSC) and a small single-copy region (SSC) of 89,345 bp and 20,021 bp, respectively. Further, the maximum likelihood phylogenetic analysis was conducted using 24 complete plastomes of the Moraceae and Cannabaceae, which supports the close relationship between Artocarpus and Morus.
Artocarpus nanchuanensis S.S. Chang is a member of the genus Artocarpus (Moraceae), which includes approximately 70 species mostly native to the hot regions of South and Southeast Asia and Oceania (William et al. Citation2017). A. nanchuanensis is unique as the northernmost species of Artocarpus, distributing at low elevations in Chongqin of SW China (Wu and Zhang Citation1989). In spite of its potential edible, medical, and industrial value, the few A. nanchuanensis communities in the field are facing a hard situation due to their rare seedlings in the understory (Sun Citation2011). In 2004, A. nanchuanensis was classified as a Critically Endangered species on the China Species Red List (Wang Citation2004). However, rare genomic data was reported about A. nanchuanensis, even the genus of Artocarpus. Here, we determined the complete chloroplast genome of A. nanchuanensis.
About 5 g leaf tissue of A. nanchuanensis was used from the specimen, which was collected from Nanchuan District, Chongqin, China (29°15′N, 107°11′E, 994.59 m) to extract genomic DNA using the modified CTAB method (Doyle and Dickson Citation1987). The voucher specimen was deposited at the Herbarium of Xishuangbanna Tropical Botanical Garden (Accession Number: XTBG-BRG-SY34421). The whole chloroplast genome was sequenced following Zhang et al. (Citation2016) and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. The contigs were aligned using the publicly available chloroplast genome of Morus indica (GenBank accession number DQ226511) and then annotated in Geneious 4.8.
The chloroplast genome of A. nanchuanensis is 160,752 bp in length, exhibiting a characteristic quadripartite structure with a pair of inverted repeats regions (IRs) of 25,693 bp, separated by a large single-copy region (LSC) and a small single-copy region (SSC) of 89,345 bp and 20,021 bp, respectively. The G + C content is 35.8%. The plastome contains a total of 122 genes (108 unique genes), among which 20 genes have one intron and three genes have two introns.
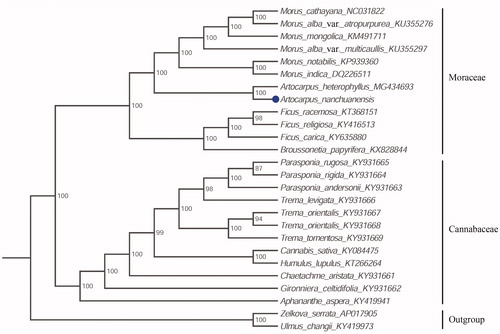
The complete plastid genome sequence of A. nanchuanensis and the other sequenced Moraceae and Cannabaceae taxa including Morus cathayana, M. alba var. atropurpurea, M. mongolica, M. alba var.multicaullis, M. notabilis, M. indica, Artocarpus heterophyllus, Ficus racemosa, F. religiosa, F. carica, Broussonetia papyrifera, Parasponia rugosa, P. rigida, P. andersonii, Trema levigata, T. orientalis, T. tomentosa, Cannabis sativa, Humulus lupulus, Chaetachme aristata, Gironniera celtidifolia and Aphananthe aspera, formed the base to perform a phylogenetic analysis, with Zelkova serrata and Ulmus changii as outgroup. A maximum likelihood analysis yielded a tree topology with 87–100% bootstrap (BS) values at each node based on GTR model in the RAxML version 8 program with 1000 bootstrap replicates (Darriba et al. Citation2012; Stamatakis Citation2014). The data supports the close relationship between Artocarpus and Morus and the robust monophyly of the Moraceae and Cannabaceae (). In the family Moraceae, Artocarpus and Morus formed a strong supported clade which was sister to the clade including Ficus and Broussonetia, consisting of previous phylogenies (Zerega et al. Citation2005; Liu et al. Citation2018).
Data archiving statement
The plastome data of the A. nanchuanensis will be submitted to Genebank of NCBI through the revision process. The accession numbers from Genebank must be supplied before the final acceptance of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 9:772.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36:715–722.
- Liu J, Niu YF, Ni SB, Liu ZY, Zheng C, Shi C. 2018. The complete chloroplast genome of Artocarpus heterophyllus (Moraceae). Mitochondrial DNA B. 3:13–14.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Sun R. 2011. Genetic resource conservation and development and utilization countermeasure of Artocarpus nanchuanensis. J Green Sci Technol. 09:20.
- Wang S. 2004. The China species Red List. Beijing: Higher Education Press.
- William EW, Gardner EM, Harris R, Chaveerach A, Pereira JT, Zerega NJC. 2017. Out of Borneo: biogeography, phylogeny and divergence date estimates of Artocarpus (Moraceae). Ann Bot. 119:611–627.
- Wu ZY, Zhang XS. 1989. Taxa Nova Nonnulla Moracearum Sinensium. Acta Bot Yunnanica. 11:24–34.
- Zerega NJC, Clement WL, Datwyler SL, Weiblen GD. 2005. Biogeography and dievrgence times in the mulberry family (Moracea). Mol Phylogenet Evol. 37:402–416.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Syst Evol. 54:219–227.