Abstract
Potato (Solanum tuberosum L.) is one of the top five staple foods in China. It has high nutritional value, adaptability, and large yield. It is the third most important food crop in the world, second only to wheat and rice. In this study, we obtained the complete chloroplast genome sequence of Potato using BGISEQ-500 sequencing. Its chloroplast genome size has 155,732 bp, containing a large single copy region (85,736 bp), a small single copy region (18,384 bp), and a pair of IR regions (25,596 bp). The overall GC contents of the chloroplast genome were 37.8%. This circular genome contains 130 annotated genes, including 85 protein-coding genes, 37 tRNAs, and eight rRNAs. Maximum likelihood phylogenetic analysis the phylogenetic relationship analysis using Neighbour-Joining (NJ) method showed that Potato (S. tuberosum L.) is much closely related to Solanum bulbocastanum. This complete chloroplast genomes can be subsequently used for the genetic breeding and useful to study the evolutionary processes in Solanaceae.
The artificial cultivation of potato (Solanum tuberosum L.) can be traced back to the southern part of Peru from about 8000 to 5000 BC (Hijmans Citation2001). It is the third most important food crop worldwide (after rice and wheat) and is number one among vegetables (Birch et al. Citation2012; Jansky et al. Citation2013). In the world, the growing importance of potato as a staple food crop can meet the demands of increasing human populations. Potato is also an important source of starch and growers face severe yield losses by pests and disease. Genomics and modern biotechnology can be used to study the molecular mechanisms of potato starch synthesis and potato molecular breeding. In this study, we obtained the complete chloroplast genome of S. tuberosum L. and explored the phylogenetic relationship with other species, which contributes to be subsequently used for the potato genetic breeding and useful to study the evolutionary in Solanaceae.
The specimen of S. tuberosum L. was isolated from College of Horticulture, Jilin Agricultural University test field in Jilin, Changchun, China (43.82N, 125.41E). The total genomic DNA of S. tuberosum L. was extracted using Plant Genomic DNA Kit (TIANGEN BIOTRCH, Beijing, CA, CHN) and stored in Jilin Agricultural University College of Horticulture (No. JAUCH01). The whole genomic DNA sequencing was performed by the BGISEQ-500 Sequencing Platform (BIG, Shenzhen, CA, CHN). The chloroplast genome was assembled with SPAdes v3.9 (Bankevich et al. Citation2012) and annotated by Blast and DOGMA (Wyman et al. Citation2004). The tRNA genes were further identified using tRNAscan-SE 2.0 (Lowe and Chan 2016). The annotated chloroplast genome was submitted to GenBank database under accession No.MH0574323.
The complete chloroplast genome of S. tuberosum L. was a circle with 155,732 bp in size, containing a large single copy region (LSC) of 85,736 bp, a small single copy region (SSC) of 18,384 bp and a pair of inverted repeat regions (IRs) of 25,596 bp. In total, 130 genes were annotated on the potato chloroplast genome, including 85 protein-coding genes (PCG), 37 transfer RNA genes (tRNA), and eight ribosomal RNA genes (rRNA). In the IR regions, a total of 16 genes were found duplicated, including 5 PCG species (rps19, rps12, rpl2, rpl23, rps7), seven tRNA species (trnI-GAU, trnL-CAA, trnV-GAC, trnI-CAU, trnA-UGC, trnR-ACG, and trnN-GUU), and four rRNA species (rrn16, rrn23, rrn4.5, and rrn5). The overall nucleotide composition is 30.7% A, 31.5% T, 19.2% C, and 18.6% G, with a total G + C content of 37.8%.
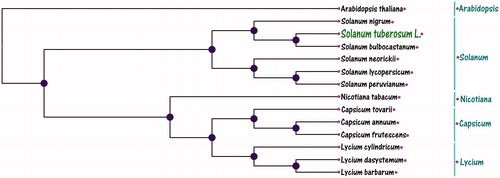
For phylogenetic analysis, we selected Potato (S. tuberosum L.) with other 13 species chloroplast genomes from GenBank to assess the relationship of S. tuberosum L. The genome-wide alignment of all chloroplast genomes was constructed using HomBlocks (Bi et al. Citation2018). The phylogenetic relationship trees were reconstructed using Neighbour-Joining (NJ) method. NJ analysis were performed using MEGA (Kumar et al. Citation2018), of which the bootstrap values were calculated using 10,000 replicates to assess node support and all the nodes were inferred with strong support by the NJ method. The final tree was represented and edited using MEGA (Kumar et al. Citation2018). As shown in the phylogenetic tree (), the chloroplast genome of showed that Potato (S. tuberosum L.) was clustered with Solanum bulbocastanum.
Figure 1. Neighbour-Joining (NJ) tree of Potato (Solanum tuberosum L.) with other 13 species chloroplast genomes. This tree was drawn without setting outgroups. All nodes exhibit above 90% bootstraps. The NCBI database accession number is AP000423.1 (Arabidopsis thaliana), NC028070.2 (Solanum nigrum), DQ347958.1 (Solanum bulbocastanu), KP117025.1 (Solanum neorickii), NC007898.3 (Solanum lycopersicum), KP117026.1 (Solanum peruvianum), NC001879.2 (Nicotiana tabacum), NC033526.1 (Capsicum tovarii), NC018552.1 (Capsicum annuum), KR078312.1 (Capsicum frutescens), NC_039651.1 (Lycium cylindricum), MG976805.1 (Lycium dasystemum), and KJ145020.1 (Lycium barbarum).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18–22.
- Birch PRJ, Bryan G, Fenton B, Gilroy EM, Hein I, Jones JT, Ankush P, Mark A, T, Lesley T, Ian K, T. 2012. Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Security. 4:477–508.
- Hijmans RJ. 2001. Global distribution of the potato crop. Am J Potato Res. 78:403–412.
- Jansky SH, Dempewolf H, Camadro EL, Simon R, Zimnoch GE, Bisognin DA, Bonierbale M. 2013. A case for crop wild relative preservation and use in potato. Crop Sci. 53:746–754.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular. Biol Evol. 35:1547–1549.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:54–57.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
