Abstract
In this study, the complete mitogenome sequence of the Favites halicora (Favites) has been sequenced by the next-generation sequence method. The overall mitogenome of F. halicora is 25.09% for A, 13.09% for C, 20.33% for G, and 41.50% for T, as well as 33.41% for low GC. The assembled mitogenome, consisting of 17,033 bp, has unique 13 protein-coding genes (PCGs), 2 transfer RNA genes, and 2 ribosomal RNA genes. The complete mitogenome provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis of stony coral phylogeny.
Keywords:
Favites halicora belongs to the family of Favites. They are widely distributes throughout the Indo-Pacific and South China Sea. Their colonies are massive, rounded or hillocky. They have corallites with thick walls and tend to become subplocoid, and paliform lobes may be developed. They are usually uniform pale yellowish or greenish-brown in color (Veron and Stafford-Smith Citation2000). There are many related species of the genus that are difficult to separate. The first establishment of F. halicora mitogenome is important for the further evolutionary and phylogenetic analysis of stony coral.
Samples (voucher no. YM01) of F. halicora were collected from Yang Meikeng sea area Daya Bay (22.5428°N, 114.5928°E) in Shenzhen, China. We used next-generation sequencing to perform whole genome sequencing according to the protocol (Niu et al. Citation2016).
The complete mitogenome of F. halicora was 17,033 bp in size (GenBank MG913991) and its overall base composition is 25.09% for A, 13.09% for C, 20.33% for G, 41.50% for T, and with 33.41% content low GC. They have circular form of complete mitogenome with about an average 205× coverage.
The protein-coding rRNA and tRNA genes of F. halicora were predicted by using DOGMA (Wyman et al. Citation2004) and MITOS (Bernt et al. Citation2013) tools and manually inspected. The complete mitogenome of F. halicora included unique 13 protein-coding genes (PCGs), 2 transfer RNA genes, and 2 ribosomal RNA genes. All genes including 13 PCGs share the start codon ATG, except for NAD2 (with TTA start codon) and COX1 (with ATT start codon). All the 13 PCGs share the stop codon TAA, except for NAD5, NAD1, NAD4, COX1 (with TAG stop codon), and ATP8 (with TGA stop codon). The longest one is NAD4 gene (1437 bp) in all PCGs, whereas the shortest is ATP8 gene (189 bp). The size of small ribosomal RNA (12S rRNA) and large ribosomal RNA (16S rRNA) gene are 911 bp and 1743 bp, respectively.
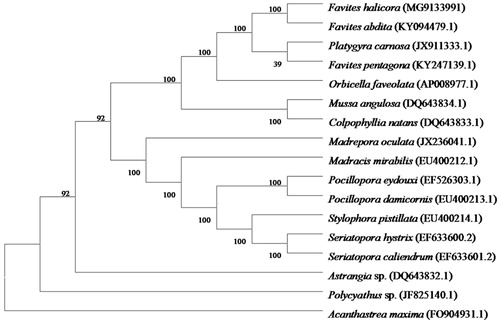
To validate the phylogenetic position of F. halicora, we used MEGA 6.0 software (Tamura et al. Citation2013) to construct a Maximum-likehood tree (with 1000 bootstrap relicates and kiumura 2-paramenter model) containing complete mitogenomes of 16 species derived from 13 different families in Scleratinia. Corallimorphus profundus derived from Corallimorphidae was used as an out-group related to F. halicora coral with high bootstrap value support (). In conclusion, the complete mitogenome of F. halicora deduced in this study provides essential and important DNA molecular data for further phylogenetic and evolutionary analysis for stony coral phylogeny.
Figure 1. Molecular phylogeny of Favites halicora and other related species in Scleractina based on complete mitogenome. The complete mitogenome is downloaded from GenBank and the phylogenic tree is constructed by the maximum-likelihood method with 1000 bootstrap replications. The gene’s accession number for tree construction is listed as follows: Favites halicora (MG9133991), Favites abdita (KY094479.1), Madracis mirabilis (EU400212.1), Stylophora pistillata (EU400214.1), Pocillopora eydouxi (EF526303.1), Pocillopora damicornis (EU400213.1), Seriatopora hystrix (EF633600.2), Seriatopora caliendrum (EF633601.2), Madrepora oculata (JX236041.1), Polycyathus sp. (JF825140.1), Mussa angulosa (DQ643834.1), Astrangia sp. (DQ643832.1), Acanthastrea maxima (FO904931.1), Colpophyllia natans (DQ643833.1), Platygyra carnosa (JX911333.1), Orbicella faveolata (AP008977.1), and Favites pentagon (KY247139.1).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Niu WT, Lin RC, Shi XF, Chen CH, Shen KN, Hsiao CD. 2016. Next generation sequencing yields the complete mitogenome of massive coral, Porites lutea (Cnidaria: Poritidae). Mitochondrial DNA Part B. 1:8–9.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Veron JEN, Stafford-Smith M. 2000. Corals of the world. Townsville: Australian Institute of Marine Science.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
