Abstract
Pteroceltis tatarinowii is a monotypic plant endemic to eastern China. It is an endangered tertiary relic and famous for Xuan paper manufacturing. In this study, we reported the Pteroceltis tatarinowii chloroplast genome with a length of 158,506 bp. In total, 132 genes were identified, including 83 protein-coding genes, 8 rRNA genes, and 33 tRNA genes. The overall GC content of this genome was 36.3%. Twenty-two species from Urticales were selected for phylogenomic analysis to determine the placement of P. tatarinowii. The complete plastome of P. tatarinowii will provide a valuable resource for further genetic conservation, phylogenomic, and evolution studies of the genus and the Ulmaceae.
Pteroceltis tatarinowii (Ulmaceae) is a tree species endemic to China with wide distribution from North to South China. This species adapts into high-calcium soil type, and can tolerate drought and barren environmental conditions. Pteroceltis tatarinowii is regarded as one of the important tree species for vegetation restoration on limestone mountains in the warm-temperate zone of eastern China (You et al. Citation2018). Moreover, because of its superior fiber texture, it is renowned as a major source for manufacturing Xuan paper, which a Chinese handmade paper has long been used by calligraphers and artists since ancient China (Mullock Citation1995). However, its geographical range is shrinking dramatically, and its population fragmentation is becoming increasingly serious because of extensive recent deforestation (Li et al. Citation2012). Here, we assembled and characterized the first complete plastome of genus Pteroceltis. It will provide potential genetic resources for further evolution and conservation studies of the genus and the other relatives in Ulmaceae.
Total DNA was extracted from fresh leaves of P. tatarinowii individual using DNA Plantzol Reagent (Invitrogen, Carlsbad, CA). The sample was collected from Shitai, Anhui, China (30°13′12″N, 117°28′48″E, Voucher No. ZSTU00686, deposited at Zhejiang University). The plastome sequences were generated using Illumina HiSeq 2500 platform. Totally, about 15.8 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome.
The full length of chloroplast genome P. tatarinowii (GenBank accession No. MK108034) is 158,506 bp (with 36.3% GC content). It consists of a large single copy region (LSC, 87,646 bp, 34.0% GC content), a small single copy region (SSC, 18,848 bp, 30.0% GC content), and two inverted repeat regions (IR, 26,006, 42.5% GC content). There were 132 genes in P. tatarinowii cp genome, including 83 protein-coding genes, 8 rRNA genes, and 33 tRNA genes. Among them, six protein-coding genes (rps19, rpl2, rpl23, ycf2, ycf15, rps7), all four rRNA genes (rrn5S, rrn4.5, rrn23, rrn16), and six tRNA genes (trnN-GTT, trnR-ACG, trnV-GAC, trnR-ACG, trnL-CAA, trnI-CAT) have two copies. Eight of these protein-coding genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhA) have two introns and one gene (ycf3) have three introns.
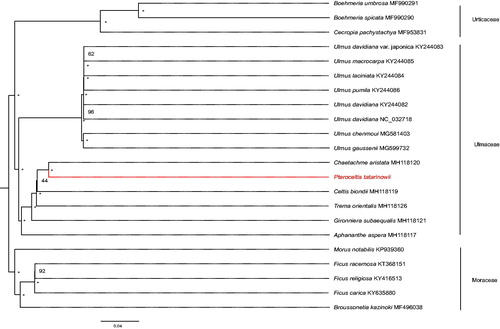
To further test the phylogentic placement of P. tatarinowii in Urticales, we selected 22 complete chloroplast genomes (3 Urticaceae species, 5 Moraceae species, and 14 species form Ulmaceae) for phylogenomic analysis. MAFFT v7.3 (Katoh and Standley Citation2013) was used to align these plastomes. Phylogenetic tree was constructed by IQTREE v1.6.7 (Nguyen et al. Citation2015) using statistical method of the maximum likelihood (ML) and model TVM + F+R3 with 5000 bootstrap replicates on the CIPRES cluster service. The result revealed that the relationship between P. tatarinowii and Chaetachme aristata was the closest within the current sampling range ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Li XH, Shao JW, Lu C, Zhang XP, Qiu YX. 2012. Chloroplast phylogeography of a temperate tree Pteroceltis tatarinowii (Ulmaceae) in China. J Systemat Evol. 50:325–333.
- Mullock H. 1995. Xuan paper. Paper Conservator. 19:23–30.
- Nguyen L, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- You HM, Fujiwara K, Tang Q. 2018. Phytosociological Study of Pteroceltis tatarinowii forest in the deciduous-forest zone of eastern China. In: Greller A, Fujiwara, Kazue M, Pedrotti F, editors, Geographical changes in vegetation and plant functional types (pp. 239–280). Cham (Zug): Springer.