Abstract
In the present study, the complete mitochondrial genome of Empoasca sp. was sequenced and assembled. The complete mitogenome of Empoasca sp. was composed of circular DNA molecules, with a total size of 15,152 bp. The base composition of this mitogenome is as follows: A (38.07%), T (40.27%), G (10.51%), and C (11.12%). The mitogenome contains 13 protein-coding genes, 2 ribosomal RNA genes (rRNA), and 22 transfer RNA (tRNA) genes. The taxonomic status of the Empoasca sp. mitogenome exhibits a closest relationship with Empoasca onukii and Empoasca vitis.
Keywords:
Empoasca is a genus of leafhoppers belonging to the family Cicadellidae, subfamily Typhlocybinae. Tea is one of the most popular beverages in the world (Ali Citation1970). It is also one of the most important cash crops in the world. China is currently the main producer and consumer of tea. Its popularity is partly attributed to its rich polyphenols, which may have a variety of health benefits, such as reducing the risk of cancer (Weisburger Citation1997). Empoasca spp. is one of the most harmful tea plant pests in China (Yang et al. Citation2017). It can seriously decrease tea yield by directly sucking juice from tender tea leaves (Yang et al. Citation2013; Xin et al. Citation2017). However, it is difficult to monitor and control due to their size and flying and jumping abilities (Shi et al. Citation2015). The mitochondrial genome of Empoasca sp. reported here will promote further understanding of the evolution, taxonomy, and population genetics of this important tea green leafhoppers.
The specimen (Empoasca sp.) was collected from a tea plantation in Ya'an, Sichuan, China (103.06 E; 30.08 N) and was stored in Sichuan Academy of Agricultural Sciences (No. Esp_1). The collected leafhoppers were initially identified as Empoasca sp. based on phenotypic characteristics (Yang et al. Citation2017). We used the E.Z.N.A.® Insect DNA Kit (Omega Bio-Tek, Norcross, GA, USA) to extract the total genomic DNA of Empoasca sp. The extracted genomic DNA was purified through a Gel Extraction Kit (Omega Bio-Tek, Norcross, GA, USA). Purified genomic DNA was stored and sequenced in the sequencing company (BGI Tech, Shenzhen, China). Sequencing libraries were constructed with purified DNA following the instructions of NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China). Whole genomic sequencing was performed by the Illumina HiSeq 2500 Platform (Illumina, SanDiego, CA). Multiple steps were used for quality control and de novo assembly of the mitogenome according to Qiang et al. (Citation2018). The SPAdes 3.9.0 software (Bankevich et al. Citation2012) was used to assemble the mitochondrial genome of Empoasca sp. Gaps among contigs were filled using MITObim V1.9 (Hahn et al. Citation2013). The complete mitochondrial genome was annotated using the MFannot tool (Valach et al. Citation2014), combined with manual corrections. tRNA genes were predicted using tRNAscan-SE v1.3.1 (Lowe and Chan Citation2016).
The total length of Empoasca sp. mitochondrial genome is 15,152 bp. This mitochondrial genome was submitted to GenBank database under accession No. MK211224. The circular mitogenome contains 13 protein-coding genes, 2 ribosomal RNA genes (rns and rnl), and 22 transfer RNA (tRNA) genes. The base composition of the genome is as follows: A (38.07%), T (40.27%), G (10.51%), and C (11.12%).
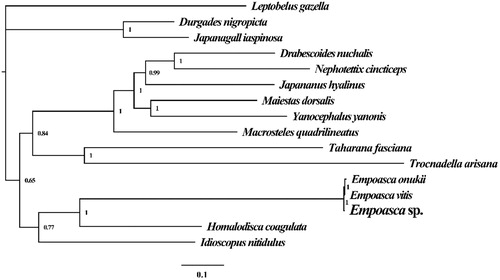
To validate the phylogenetic position of Empoasca sp., we construct the phylogenetic trees of 16 closely related species based on the nucleotide sequences of the 13 core PCGs (atp6, atp8, cytb, cox1, cox2, cox3, ND1, ND2, ND3, ND4, ND4L, ND5, and ND6), in addition to the rns and rnl mitochondrial genes. Bayesian inference (BI) phylogenetic methods were used to construct phylogenetic trees using the combined gene datasets with MrBayes v3.2.6 (Ronquist et al. Citation2012). Bayesian posterior probabilities (BPP) were calculated to assess node support. As shown in the phylogenetic tree (), the taxonomic status of the Empoasca sp. based on combined mitochondrial gene dataset exhibits a closest relationship with Empoasca onukii (Liu et al. Citation2017) and Empoasca vitis (Zhou et al. Citation2016).
Figure 1. Molecular phylogenies of 16 species based on the Bayesian inference analysis of the combined mitochondrial gene set (13 core protein-coding genes + 2 rRNA genes). Node support values are Bayesian posterior probabilities (BPP). Mitogenome accession numbers used in this phylogeny analysis: Empoasca onukii (NC_037210), Trocnadella arisana (NC_036480), Japananus hyalinus (NC_036298), Maiestas dorsalis (NC_036296), Yanocephalus yanonis (NC_036131), Taharana fasciana (NC_036015), Japanagallia spinosa (NC_035685), Durgades nigropicta (NC_035684), Macrosteles quadrilineatus (NC_034781), Idioscopus nitidulus (NC_029203), Drabescoides nuchalis (NC_028154), Nephotettix cincticeps (NC_026977), Empoasca vitis (NC_024838), Leptobelus gazelle (NC_023219), and Homalodisca vitripennis (NC_006899).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ali L. 1970. The regulation of trade in tea. J World Trade Law. 4:565–485.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44:W54–W57.
- Liu JH, Sun CY, Long J, Guo JJ. 2017. Complete mitogenome of tea green leafhopper, Empoasca onukii (Hemiptera: Cicadellidae) from Anshun, Guizhou Province in China. Mitochondrial DNA Part B. 2:808–809.
- Qiang L, Mei Y, Cheng C, Chuan X, Xin J, Zhigang P, Wenli H. 2018. Characterization and phylogenetic analysis of the complete mitochondrial genome of the medicinal fungus Laetiporus sulphureus. Sci Rep. 8:9104.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Shi LQ, Zeng ZH, Huang HS, Zhou YM, Vasseur L, You MS. 2015. Identification of Empoasca onukii (Hemiptera: Cicadellidae) and monitoring of its populations in the tea plantations of South China. J Econ Entomol. 108:1025–1033.
- Valach M, Burger G, Gray MW, Lang BF. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42:13764–13777.
- Weisburger JH. 1997. Tea and health: a historical perspective. Cancer Lett. 114:315–317.
- Xin ZJ, Li XW, Bian L, Sun XL. 2017. Tea green leafhopper, Empoasca vitis, chooses suitable host plants by detecting the emission level of (3Z)-hexenyl acetate. Bull Entomol Res. 107:77–84.
- Yang CS, Li G, Yang Z, Guan F, Chen A, Ju J. 2013. Cancer prevention by tocopherols and tea polyphenols. Cancer Lett. 334:79–85.
- Yang TB, Liu J, Yuan LY, Zhang Y, Li DQ, Agnarsson I, Chen J. 2017. Molecular identification of spiders preying on Empoasca vitis in a tea plantation. Sci Rep. 7:7784.
- Zhou N, Wang M, Cui L, Chen X, Han B. 2016. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae). Mitochondrial DNA A. 27:1052–1053.
