Abstract
The complete genome of Vaccinium oldhamii plastid was sequenced and analyzed in this study. The V. oldhamii plastid a circular genome of 173,245 bp with 33 genes, including 88 protein, 37 tRNA, and eight rRNA genes. Phylogenetic analysis showed that species of Vaccinium were the first branching taxon In Ericaceae.
Vaccinium (Ericaceae) comprises 400–450 species of deciduous or evergreen shrubs that grow almost worldwide. Among these, Vaccinium oldhamii Miq. is a deciduous shrub of 2–3 m height that is found in Korea, China, and Japan. The fruit of some wild Vaccinium species have been collected and used as food by humans for centuries (Hirai et al. Citation2010). In particular, antioxidants present in blueberries are effective in the prevention of cancer, heart disease, and vascular and neurodegenerative diseases. The fruit of V. oldhamii has been found to be most effective in inhibiting the growth of leukemia cells (Tsuda et al. Citation2013). Because of economic, agricultural, and medical importance, this study investigated and assembled the complete chloroplast sequences of pure wood using Ion torrent sequencing data. We propose a molecular phylogenetic relationship in relation to the chloroplasts of other plants of Ericales.
Plant material (Voucher specimen: WTFRC 10015918) was collected from Sillye-ri, Namwon-eup, Seogwipo-si, Jeju (33°18′ N, 126°37′ E), and genomic DNA was isolated from fresh leaves using a Plasmid SV mini kit (GeneAll, Seoul, Korea). All samples collected stored in a DNA bank in the Forest Genetic Resources Department (Suwon, Korea). Whole genome sequencing was conducted using Ion Torrent sequencing platform (Life Technologies, Carlsbad, CA). Filtered sequences were assembled with the reference sequence of Vaccinium macrocarpon (GenBank: JQ757046). The sequenced fragments were assembled using Geneious R10 (Biomatters Ltd, Auckland, New Zealand; Kearse et al. Citation2012). Annotation was performed using DOGMA (http://dogma.ccbb.utexas.edu/) and BLAST searches. All tRNA sequences were confirmed using the web-based tool tRNAScan-SE (Schattner et al. Citation2005) with default settings to corroborate tRNA boundaries identified using Geneious. The maximum likelihood (ML) tree searches and ML bootstrap searches were performed using RAxML BlackBox web-server (https://raxml-ng.vital-it.ch/#/, Stamatakis et al. Citation2008) with 79 protein-coding genes from the plastid genomes of nine species. The RAxML analyses were run with rapid bootstrap analysis using a random starting tree and 100 ML bootstrap replicates.
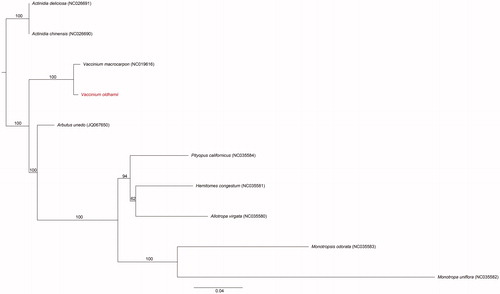
The plastome of V. oldhamii was determined to be a double-stranded, circular DNA of 173,245 bp containing two inverted repeat regions (IRs) of 32,340 bp each that were separated by large single-copy (LSC) and small single-copy (SSC) regions of 105,498 and 3,067 bp, respectively (NCBI accession no. MK049537). The genome contained 133 genes, including 88 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The 11 protein-coding genes, eight tRNA genes, and four rRNA genes were duplicated in the IR. The overall GC content was 36.8% (LSC, 35.8%; SSC, 29.2%; IRs, 38.7%). The ML phylogenetic tree constructed from the genomes of 10 species of Ericaceae (including outgroup: Actinidiaceae) showed that species of Vaccinium were the first branching taxon in Ericaceae ().
Disclosure statement
The authors report no declaration of interest. The authors alone are responsible for the content and writing of this article.
References
- Hirai M, Yoshimura S, Ohsako T, Kubo N. 2010. Genetic diversity and phylogenetic relationships of the endangered species Vaccinium sieboldii and Vaccinium ciliatum (Ericaceae). Plant Syst Evol. 287:75–84.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57:758–771.
- Tsuda H, Kunitake H, Kawasaki-Takaki R, Nishiyama K, Yamasaki M, Komatsu H, Yukizaki C. 2013. Antioxidant activities and anti-cancer cell proliferation properties of Natsuhaze (Vaccinium oldhamii Miq.), Shashanbo (V. bracteatum Thunb.) and Blueberry cultivars. Plants. 2:57–71.