Abstract
In the present study, a complete mitochondrial genome of the moon crab species, Matuta planipes was sequenced and analyzed. This is the second complete mitochondrial genome record for the species and the first record from South Korea. The genome structure and gene orientation are identical but the protein-coding gene sizes are slightly different in comparison with the previous record. Additionally, the phylogenetic relationship of M. planipes was investigated using protein-coding genes of the complete mitochondrial genome. The present study suggests that the closest species to M. planipes is Ashtoret lunaris and they belong to the family Matutidae. This study provides additional data for the family Matutidae phylogeny.
Matutidae is the family of the moon crabs which are easily distinguishable with their paddle-shaped walking legs (Tan et al. Citation2016). This special anatomy allows them not only swimming but also digging the sand (Lin et al. Citation2018). The family Matutidae contains 15 species placed in four genera (WORMS Citation2018). Although number of species in the family is limited, the phylogenetic relationships are still contradictory (Lin et al. Citation2018). There are speculations on the relationship of Matutidae within the superfamily Calappoidea (Ng et al. Citation2008; Silambarasan et al. Citation2014). Among them, only Ashtoret lunaris from Sri Lanka and Matuta planipes from China have complete mitochondrial genome records (Tan et al. Citation2016; Lin et al. Citation2018). In this study, we are providing the second complete mitochondrial genome sequence of the flower moon crab M. planipes from South Korea.
The M. planipes specimen was collected from Eurwangni Beach, Incheon, South Korea (37°26'52″ N, 126°22'15″ E) on September 2016 and deposited in Department of Biotechnology, Sangmyung University, Korea University (SM00030). After species identification by DNA barcoding, the total genomic DNA was isolated from the walking leg. The methods for mitochondrial genome sequencing and phylogeny analysis were described in the previous study (Karagozlu et al. Citation2016).
The mitochondrial genome of M. planipes is 15,751 bp in length (GenBank accession number: MK281334). The mitochondrial genome consists of 13 protein-coding, two rRNAs, and 22 tRNAs genes. Among these genes, four protein-coding genes, two rRNA, and eight tRNAs encoded in minor strand while the remaining 23 genes encoded in major strand. The nucleotide composition of the mitochondrial genome is 34.9% A, 18.2% C, 11.1% G, and 35.8%. There are 11 overlapping regions in the genome show length variation ranging from 1 to 20 bp while there are 12 intergenic sequences show length variation ranging from 1 to 38 bp. The longest intergenic sequence is between tRNA-Phe and nd5 genes, and the longest overlapping region is between cox2 and tRNA-Lys. Likewise the other record, there is a control region in between 12s rRNA and tRNA-Ile which is 911 bp in length. This area is 925 bp long in the Chinese record. This region is the least similar region in the M. planipes records. The similarity between Korean and Chinese records for this region is 85.5%. Likewise size differences in the control regions, there are some size differences in protein-coding genes too. In comparison of the two records, cox1 (5 bp), cox2 (20 bp), cytb (6 bp), nd3 (2 bp), nd4 (3bp), nd4l (6 bp), and nd5 (3 bp) genes are longer in the Korean record. The reason for the size difference is the location of the stop codon.
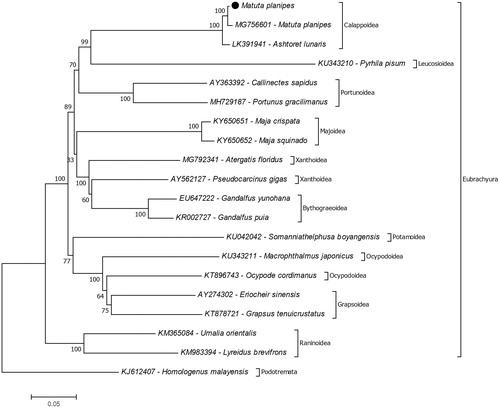
The reconstruction of the phylogenetic tree suggests that the closest species to M. planipes is Ashtoret lunaris and they belong to the monophyletic superfamily Calappoidea (). Unfortunately, there is not enough information on the complete mitochondrial genome of the moon crabs to confirm their phylogenetic relationships. For further studies, complete mitochondrial genome data records should increase to investigate molecular phylogeny.
Figure 1. Molecular phylogeny of Matuta planipes in the section Eubrachyura. The species from the section Podotremata represents outgroup. The phylogeny tree reconstructed due to protein-coding genes of mitochondrial genome with maximum likelihood statistical method using MEGA 7 software. mtREV with Freqs (+F) model used for amino acid substitution and bootstrap method replicated 1000 times for the test of phylogeny. The data provided in the present study indicated with black dot.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Karagozlu MZ, Sung JM, Lee JH, Kwon T, Kim CB. 2016. Complete mitochondrial genome sequences and phylogenetic relationship of Elysia ornata (Swainson, 1840) (Mollusca, Gastropoda, Heterobranchia, Sacoglossa). Mitochondrial DNA B Res. 1:230–232.
- Lin F, Tan H, Fazhan H, Xie Z, Guan M, Shi X, Ma H. 2018. The complete mitochondrial genome and phylogenetic analysis of Matuta planipes (Decapoda: Brachyura: Matutidae). Mitochondrial DNA B Res. 3:157–158.
- Ng PKL, Guinot D, Davie P. 2008. Systema Brachyurorum: part 1. An annotated checklist of extant brachyuran crabs of the world. Raff Bull Zool. 17:1–286.
- Silambarasan K, Velmurugan K, Rajalakshmi E, Anithajoice A. 2014. Redescription and new distributional records of Matuta planipes (Fabricius, 1798) (Crustacea; Decapoda; Matutidae) from Chennai Coast, Tamil Nadu. Arthropods. 3:43–47.
- Tan MH, Gan HM, Lee YP, Austin CM. 2016. The complete mitogenome of the moon crab Ashtoret lunaris (Forskal, 1775), (Crustacea; Decapoda; Matutidae). Mitochondrial DNA A. 27:1313–1314.
- WoRMS Editorial Board. 2018. World Register of Marine Species. Available from http://www.marinespecies.org at VLIZ. [accessed 2018 Nov 18].
