Abstract
Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier is one of the common weeds as well as a model organism of a basal group of land plants. To understand the genetic diversity of M. polymorpha subsp. ruderalis based on geographical distribution, we completed chloroplast genome of KBDI00084 isolated from Mt. Cheonma in Korea. Its length is 120,304 bp, consisting four subregions: 80,706 bp of large single copy (LSC) and 19,788 bp of small single copy (SSC) regions are separated by 9,905 bp of inverted repeats (IRs). 134 genes (90 protein-coding genes, eight rRNAs, and 36 tRNAs) were annotated successfully. The overall GC content of the chloroplast genome is 29.0% and those in the LSC, SSC, and IR regions are 26.6%, 25.1%, and 42.5%, respectively. Only four single nucleotide polymorphisms (SNPs) are identified in KBDI00084, while that of Poland isolate presents 69 SNPs and 660 insertions and deletions. Phylogenetic trees show that four chloroplast genomes of M. polymorpha subsp. ruderalis are clustered into one clade. Taken together, the reason of low genetic diversity between Korea and Japan may be affected by human activities on M. polymorha subsp. ruderalis.
The liverwort Marchantia polymorpha L. is one of the common weeds that grows in nursery crop production (Altland et al. Citation2007) and distributed widely around the world from tropical to arctic climates (Shimamura Citation2016). Marchantia polymorpha is one of the model organisms representing a basal group of land plants, so whole genome of Marchantia polymorpha subsp. ruderalis was sequenced including chloroplast genome (Bowman et al. Citation2017). Unlike the other two subspecies of M. polymorpha, M. polymorpha subsp. ruderalis is a weedy species usually found in human habitats (Bischler-Causse and Boisselier-Dubayle Citation1991). It is important to understand the effects of human activities on genetic diversity based on the geographical distribution because human activities have affected ecological environments in various ways (Lenzen et al. Citation2012).
We collected M. polymorpha subsp. ruderalis isolated in Mt. Cheonma, Korea (InfoBoss Cyber Herbarium (IN); Kwon W., IB-50002). Total DNA was extracted from fresh thalli of M. polymorpha subsp. ruderalis using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeq4000 at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008). All bases were confirmed by alignment generated by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation with M. polymorpha subsp. ruderalis Kitashirakawa-2 chloroplast genome (reference genome; NC_037507).
The length of chloroplast genome of Korean M. polymorpha subsp. ruderalis, named as KBDI00084 (Genbank accession is MK202952), is 120,304 bp, which is similar to the reference genome. It consists of four subregions: 80,706 bp of large single copy (LSC) and 19,788 bp of small single copy (SSC) regions are separated by 9,905 bp of inverted repeats (IRs). It contains 134 genes (90 protein-coding genes, 8 rRNAs, and 36 tRNAs) and overall GC ratio is 29.0% and those in LSC, SSC, and IR regions are 26.6%, 25.1%, and 42.5%, respectively.
We identified four single nucleotide polymorphisms (SNPs) and no insertions and deletions (INDELs) on KBDI00084 against the reference genome. In addition, chloroplast genome of another isolate, Takaragaike-1 (Takenaka et al. Citation2000), used for sequencing its Y chromosome (Yamato et al. Citation2007), was assembled from raw reads (SRR896230) presenting no variation against the reference chloroplast genome. It is a striking result because Korea and Japan are geographically isolated enough to be diverged from M. polymorpha subsp. ruderalis. In addition, Polish isolate (MG762001) chloroplast contains 69 SNPs and 660 INDELs against the reference genome, supporting that M. polymorpha subsp. ruderalis chloroplast has enough variations among different isolates from different regions.
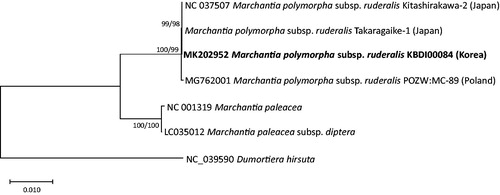
Six Marchantia (Shimda and Sugiuro Citation1991; Bowman et al. Citation2017) and one Dumortiera (Kwon et al. Citation2019) complete chloroplast genomes were analyzed for constructing maximum likelihood and neighbor joining phylogenetic trees utilizing MAFFT 7.388 (Katoh and Standley Citation2013) and MEGA X (Kumar et al. Citation2018). The trees present that M. polymorpha subsp. ruderalis from three countries are clustered in one clade (). Taken together, the reason of low genetic diversity between Korea and Japan may be affected by human activities on M. polymorha subsp. ruderalis, which is for next research.
Figure 1. Maximum likelihood (bootstrap repeat is 1000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees of Marchantiaceae based on seven complete chloroplast genomes including one outgroup species: M. polymorpha subsp. ruderalis (MK202952 in this study, NC_037507, Takaragaike-1, and MG762001), Marchantia paleacea (NC_001319), Marchantia paleacea subsp. diptera (LC035012), and Dumortiera hirsuta (NC_039590) as an outgroup species. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Altland JE, Wehtje G, Gilliam CH, Miller ME. 2007. Liverwort (Marchantia polymorpha) control with quinoclamine. Weed Technol. 21:483–488.
- Bischler-Causse H, Boisselier-Dubayle M. 1991. Lectotypification of Marchantia polymorpha L. J Bryol. 16:361–365.
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171:287–304. e215.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Kwon W,Kim Y,Park J. 2019. The complete chloroplast genome sequence of Dumortiera hirsuta (Sw.) Nees (Marchantiophyta, Dumortieraceae). Mitochondrial DNA Part B. 4: 318–319.
- Lenzen M, Moran D, Kanemoto K, Foran B, Lobefaro L, Geschke A. 2012. International trade drives biodiversity threats in developing nations. Nature. 486:109.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Shimamura M. 2016. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 57:230–256.
- Shimda H, Sugiuro M. 1991. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 19:983–995.
- Takenaka M, Yamaoka S, Hanajiri T, Shimizu-Ueda Y, Yamato KT, Fukuzawa H, Ohyama K. 2000. Direct transformation and plant regeneration of the haploid liverwort Marchantia polymorpha L. Transgenic Res. 9:179–185.
- Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, Bando H, Yodoya K, Hayashi K, Bando T, et al. 2007. Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc Natl Acad Sci. 104:6472–6477.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
