Abstract
The aromatic plant immortelle contains valuable bioactive molecules and a unique fragrance. These key factors contribute to its attractiveness and implementation in agricultural ecosystems as well as applications in the pharmaceutical and cosmetic industries. In this study, we have characterized the complete chloroplast genome sequence of Helichrysum italicum subsp. italicum from the North Adriatic region. The total genome size was 152,431 bp in length, containing 85 protein-coding genes, 36 transfer RNA genes, 8 ribosomal RNA genes, and 2 partial genes (ycf1 and rps19). Based on the available chloroplast genomes from Gnaphalieae tribe, the closest relationship was identified with Anaphalis sinica.
Helichrysum italicum is one of the members of the Helichrysum italicum complex (Asteraceae family, Gnaphalieae tribe, Stoechadina (DC.) Gren. & Godr. section), which is widely distributed in the Mediterranean region. The species H. italicum is further divided into four subspecies italicum, siculum, microphyllum, tyrrhenicum based on morphological features, geographical distribution and identified genetic variation (Herrando-Moraira et al. Citation2016). Overlapping of subsp. and their morphological plasticity often prevents a reliable classification of subspecies, supporting the necessity for new high-informative DNA markers. We report here the complete chloroplast genome sequence of Helichrysum italicum subsp. italicum as a source for the development of new molecular markers for various genetic studies.
Leaves were collected from an individual plant growing in Črišnjeva, Croatia (45°14′56.4″N, 14°34′47.7″E). The original DNA sample is stored in the laboratory of the University of Primorska, Slovenia (specimen number Hii16072018UP). Total genomic DNA was extracted with the CTAB protocol (Japelaghi et al. Citation2011).
The NGS whole genome shotgun DNA library was prepared with the Ion Plus Fragment Library kit using size selection with E-gelTM system (Thermo Fisher Scientific, Waltham, MA, USA). The library was amplified on an Ion OneTouchTM 2 System with Ion 520TM & 530TM OT2 Kit (400 bp option) and sequenced on an Ion S5TM System using Ion 530TM chip (Thermo Fisher Scientific, Waltham, MA, USA).
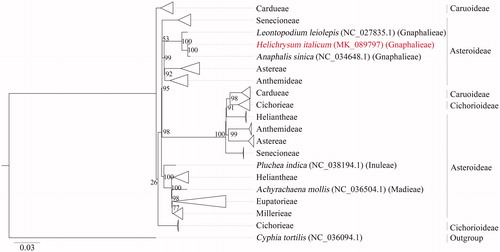
In total, 17,025,150 high-quality sequences were obtained. The chloroplast genome was assembled employing the Fast-Plast v1.2.8 pipeline (https://github.com/mrmckain/Fast-Plast) (McKain and Wilson Citation2018) and further annotated with the GeSeq web application (Tillich et al. Citation2017). CLC Genomics Workbench 11 (QIAGEN, Valencia, CA, USA) was used for manual curation of automated annotations. LSC, SSC, and IR regions were additionally confirmed with an IRSCOPE tool (Amiryousefi et al. Citation2018). The annotated chloroplast genome was submitted to the GenBank database with assigned Accession No. MK089797. The phylogenetic relationship of H. italicum was conducted based on the 166 complete chloroplast genomes of species from the Asteraceae family. The complete chloroplast genome sequences were aligned with MAFFT v7.2 software (Katoh and Standley Citation2013) and the evolutionary analyses were conducted with RAxML v8.2.10 (Stamatakis Citation2014) on the CIPRES Science Gateway (Miller et al. Citation2010). The GTRCAT model with ‘Let RAxML halt bootstrapping automatically’ was used. The phylogenetic tree was visualized using FigTree v1.4.4 (FigTree, Citation2018).
The complete chloroplast genome of H. italicum was 152,431 bp in length. The chloroplast genome contained 131 genes, including 85 protein-coding genes, 36 transfer RNA genes, 8 ribosomal RNA genes, and 2 partial genes (ycf1 and rps19). The same number of genes was previously determined by Dong-Hyuk et al. (Citation2017) in Anaphalis sinica. The lengths of LSC, IR regions, and SSC were 84.445 bp, 24.773 bp, and 18.440 bp, respectively. The overall base composition of the chloroplast was 31.3% A, 18.3% C, 18.8% G, and 31.5% T.
Two plant species from the Gnaphalieae tribe, Anaphalis sinica, and Leontopodium leiolepis, formed a common group (), whereas the closest relationship was with Anaphalis sinica. A close relationship of Anaphalis with Helichrysum genus was previously identified by Nie et al. (Citation2013).
Figure 1. The phylogenetic tree constructed with 166 chloroplast genomes of different species (deposited in GenBank at the time of analysis) from Asteraceae family. Species Cyphia tortilis (NC_036094.1) was used as an outgroup. The clades with samples from the same tribe were collapsed (indicated with triangles) due to high number of the samples. Node labels indicate bootstrap values.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Amiryousefi A, Hyvönen J, Poczai P. 2018. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 34:3030–3031.
- Dong-Hyuk L, Won-Bum C, Byoung-Hee C, Jung-Hyun L. 2017. Characterization of two complete chloroplast genomes in the tribe Gnaphalieae (Asteraceae): Gene loss or pseudogenization of trnT-GGU and implications for phylogenetic relationships. Hortic Sci Technol. 35:769–783.
- FigTree - Molecular Evolution, Phylogenetics and Epidemiology. 2018. [accessed 2018 Nov 26]. http://tree.bio.ed.ac.uk/software/figtree/.
- Herrando-Moraira S, Blanco-Moreno JM, Sáez L, Galbany-Casals M. 2016. Re-evaluation of the Helichrysum italicum complex (Compositae: Gnaphalieae): a new species from Majorca (Balearic Islands). Collect Bot. 35:e009.
- Japelaghi R, Haddad R, Garoosi G-A. 2011. Rapid and efficient isolation of high quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Mol Biotechnol. 49:129–137.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- McKain MR, Wilson M. 2018. Fast-plast: rapid de novo assembly and finishing for whole chloroplast genomes. mrmckain/Fast-Plast: Fast-Plast v.1.2.8. Zenodo.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14; New Orleans, Louisiana: Institute of Electrical and Electronics Engineers (IEEE); p. 1–8.
- Nie Z-L, Funk V, Sun H, Deng T, Meng Y, Wen J. 2013. Molecular phylogeny of Anaphalis (Asteraceae, Gnaphalieae) with biogeographic implications in the Northern Hemisphere. J Plant Res. 126:17–32.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
