Abstract
In this study, the complete mitochondrial genome of Cosmoscarta dorsimacula (Hemiptera, Cicadomorpha, Cercopoidea, Cercopidae) was firstly determined. The complete genome is 15,462 bp in length. It contains 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and a control region (A + T-rich region). The gene organization, nucleotide composition, and codon usage are similar to other Cercopoidea mitogenomes. The overall nucleotide composition was 44.61% A, 34.01% T, 12.53% C, and 8.85% G, respectively. Phylogenetic analysis highly supported that C. dorsimacula showed a close relationship with C. bispecularis. The measure of complete mitogenome sequence of C. dorsimacula will provide fundamental data for the phylogenetic and biogeographic studies of the Cercopoidea and Hemiptera.
Cosmoscarta dorsimacula is a common pest on glutinous rice yam (Dioscoreaalata L.).Glutinous rice yam is a specific local variety in Wenzhou city, and the yield of yam is affected by C. dorsimacula. C. dorsimacula emerges one generation each year in Zhejiang province, China. Most of them spend the winter in the host stalk and hatch in April next year. Both adult and nymph inhabit glutinous rice yam and so on, mainly sucking its juice. So it is important to sequence and annotate the mitochondrial genome (mito-genome) of C. dorsimacula. Here, we report the complete mitogenome sequence of C. dorsimacula by next-generation sequencing (NGS). The annotated mitochondrial DNA (mtDNA) sequence has been deposited in GenBank under accession number MK256929.
In this study, the specimens were collected from Wenzhou, Zhejiang province(27°49′3″N, 120°7′6″E) of China in September 2017, and were deposited in the insect collection bank of Wenzhou Vocational College of Science and Technology (Wenzhou, China).
The specimens were initially preserved in 100% ethanol and then stored at 4 °C prior to DNA extraction. Total genomic DNA was extracted from the individual adult specimen using a DNeasy tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. One library was constructed with VAHTS™ Universal DNA Library Prep Kit for Illumina®, and sequenced with lllumina HiSeq X Ten sequencer (150 bp pared-end) by Novogene (Beijing, China).
The complete mitochondrial genome of C. dorsimacula was a circular molecule with 15,462 bp in length (GenBank Accession number: MK256929). The mitogenome of C. dorsimacula contained 2 rRNA genes, 13 protein-coding genes (PCGs), 22 tRNA genes and a control region. The overall nucleotide composition was 44.61% A, 34.01% T, 12.53% C, 8.85% G, with a slight AT bias of 78.62%. Seven PCGs initiation codons were ATG, four PCGs initiation codons were ATA, two PCGs began with ATT, different from C. bispecularis (ND2 and ND3 with ATA, ND5 with ATT) (Yang et al. Citation2016). Correspondingly, six PCGs stopped with the complete termination codon TAA, two PCGs stopped with the complete termination codon TAG, while the rest of PCGs ended with an incomplete termination codon T (COX1, COX2, ND3, ND4, ND5), which was a little different from C. bispecularis (ND3 with TAG) (Yang et al. Citation2016). Moreover, the 22 tRNA genes ranged in size from 61 bp (tRNAGly) to 74 bp (tRNAIle). The two rRNA genes of C. dorsimacula were determined by sequence alignment with other species. The lrRNA is located between the tRNALeu (TAG) and tRNAVal with a 1246 bp in length, while the srRNA is located between the tRNAVal and the control region with a 773 bp in length.
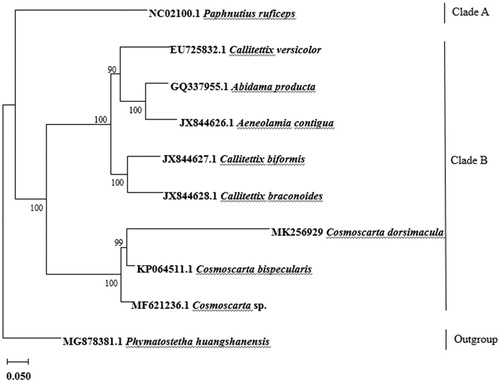
To confirm the phylogenetic relationships between C. dorsimacula and other Cercopidae, phylogenetic analysis was performed on the concatenated dataset of 13 PCGs using maximum likelihood (ML) method that produced the ML tree (Liu and Liang Citation2013; Liu et al. Citation2014; Su and Liang Citation2018) (). Phymatostetha huangshanensis (Cercopoidea, Cercopidae) was defined as an outgroup. The other nine species were divided into two clades. Paphnutius ruficeps was clustered into clade A, and the rest of species were clustered into clade B. C. dorsimacula, C. bispecularis, and Cosmoscarta sp. were grouped in one clade, suggested the close relationship of these species, and further confirmed that C. dorsimacula belongs to the subfamily Cercopinae. C. dorsimacula was evolutionarily closest to C. bispecularis, and Cosmoscarta sp. was a sister of C. dorsimacula (). Each of these relationships had 100% bootstrap support.
Acknowledgements
We are grateful to Dr. Song Shengnan from Oil Crops Research Institute, Chinese Academy of Agricultural Science (Wuhan), Dr. Tang Pu and Zheng Boyingfrom Zhejiang University for their help in this study.
Disclosure statement
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Liu J, Bu CP, Wipfler B, Liang AP. 2014. Comparative analysis of the mitochondrial genomes of Callitettixini Spittlebugs (Hemiptera: Cercopidae) confirms the overall highevolutionary speed of the AT-rich region but reveals the presenceof short conservative elements at the tribal level. PLoS One. 9:e109140.
- Liu J, Liang AP. 2013. The complete mitochondrial genome of spittlebug Paphnutius ruficeps (lnsecta: Hemiptera: Cercopidae)with a fairly short putative control region. Acta Biochimica et Biophysica Sinica. 45:309–319.
- Su TJ, Liang AP. 2018. Characterization of the complete mitochondrial genome of Phymatostetha huangshanensis (Hemiptera: Cercopidae) and phylogenetic analysis. Int J Biol Macromol. 119:60–69.
- Yang H, Liu J, Liang AP. 2016. The complete mitochondrial genome of Cosmoscarata bispecularis (Hemiptera, Cicadomorpha, Cercopoidea, Cercopidae). Mitochond DNA Part A. 27:3957–3958.