Abstract
Illicium anisatum L. is a neighbor species of Illicium verum used for making anti-virus vaccine. Here, we completed the chloroplast genome of I. anisatum. Its length is 142,747 bp long and has four subregions: 100,833 bp of large single copy (LSC) and 20,227 bp of small single copy (SSC) regions are separated by 10,844 bp of inverted repeat (IR) including 123 genes (81 protein-coding genes, 8 rRNAs, and 34 tRNAs). The overall GC content of this chloroplast genome is 39.2% and those in the LSC, SSC, and IR regions are 38.1%, 33.9%, and 49.3%, respectively. Twenty-one single nucleotide polymorphisms (SNPs) and 114 insertion and deletions (INDELs) are identified against I. anisatum originated from Green Farmacy Garden. Interestingly, 12 INDEL clusters, of which length ranges from 2 bp to 21 bp are identified, which is like the case of Duchesnea chrysantha. Phylogenetic trees show that five Illicium chloroplast genomes form Illicium clade, clearly.
Illicium L. is a sole genus of Illiciaceae recognized as an independent family until APG II system (Angiosperm Phylogeny Group Citation2003) but it was merged with Schisandraceae since APG III (Angiosperm Phylogeny Group Citation2009; Chase et al. Citation2016). Illicium covers at least 42 species (Smith Citation1947), which is a notorious genus of which morphological characters with causing severe misidentification (Morris et al. Citation2007). Illicium anisatum L. distributed in Korea, Japan, and Taiwan is a neighbor species of I. verum containing shikimic acid, used for anti-virus vaccine (Edmonds and Payne Citation2005). Illicium anisatum cannot be easily distinguished with I. verum morphologically (Morris et al. Citation2007), causing severe symptoms such as epilepsy, hallucinations, and nausea induced by anisatin, shikimin, sikimitoxin, and neoanisatin. (Lane et al. Citation1952; Yamada et al. Citation1968; Herrmann and Weaver Citation1999; Parthasarathy et al. Citation2008; Shen et al. Citation2012). Several techniques to distinguish two species have been established (Fritz et al. Citation2008; Shen et al. Citation2012) to avoid this problem.
Total DNA of I. anisatum isolated from Gujwa-eup, Jeju-si, Jeju-do, Korea (Voucher in InfoBoss Cyber Herbarium (IN); Y. Kim, IB-00585) was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome was sequenced using HiSeqX at Macrogen Inc., Korea, and de novo assembly and confirmation were performed by Velvet 1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser 1.12 (Zhao et al. Citation2011), BWA 0.7.17 (Li Citation2013), and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd., Auckland, New Zealand) was used for annotation based on I. anisatum chloroplast complete genome (NC_034703) from Green Farmacy Garden (MD, USA)
The chloroplast genome of I. anisatum (Genbank accession is MK358442) is 142,747 bp (GC ratio is 39.2%) and has four subregions: 100,833 bp of large single copy (LSC; 38.1%) and 20,227 bp of small single copy (SSC; 33.9%) regions are separated by 10,844 bp of inverted repeat (IR; 49.3%). It contains 123 genes (81 protein-coding genes, 8 rRNAs, and 34 tRNAs); 10 genes (one protein-coding gene, 4 rRNAs, and 5 tRNAs) are duplicated in IR regions, smaller than those of other Illicium species.
By comparing with Green Farmacy Garden chloroplast, 21 single nucleotide polymorphisms (SNPs) and 114 insertion and deletions (INDELs) are identified. Two SNPs are in ndhA: one is synonymous and the other is nonsynonymous. Rest are in intergenic regions. Twelve INDEL groups are identified ranging from 2 bp to 21 bp. It is like two Duchesnea chrysantha chloroplast genomes containing three INDEL groups (one is 19 bp and two are 20 bp; Park et al., 2019).
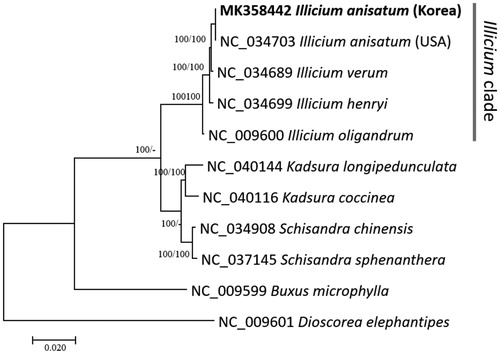
Five Illicium and six chloroplast genomes from Schisandraceae were used for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenic trees using MEGA X (Kumar et al. Citation2018) after aligning whole chloroplast genomes using MAFFT 7.388 (Katoh and Standley Citation2013). Phylogenetic trees show that five Illicium chloroplasts are clustered in one clade and I. verum is clearly separated from I. anisatum with high bootstrap support (), indicating molecular markers can be another technique to distinguish.
Figure 1. Neighborjoining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of five Illicium and six Schisandraceae chloroplast genomes: Illicium anisatum (MK358442 in this study and NC_034703), Illicium verum (NC_034689), Illicium henryi (NC_034699), Illicium oligandrum (NC_009600), Kadsura longipedunculata (NC_040144), Kadsura coccinea (NC_040116), Schisandra chinensis (NC_034908), Schisandra sphenanthera (NC_037145), Buxus microphylla (NC_009599), and Dioscorea elephantipes (NC_009601). Phylogenetic tree was drawn based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Angiosperm Phylogeny Group. 2003. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linnean Soc. 141:399–436.
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linnean Soc. 161:105–121.
- Chase MW, Christenhusz M, Fay M, Byng J, Judd WS, Soltis D, Mabberley D, Sennikov A, Soltis PS, Stevens PF. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linnean Soc. 181:1–20.
- Edmonds M, Payne R. 2005. Isolation of shikimic acid from star aniseed. J Chem Educ. 82:599.
- Fritz E, Ölzant SM, Länger R. 2008. Illicium verum Hook. f. and Illicium anisatum L.: anatomical characters and their value for differentiation. Sci Pharm. 76:65–76.
- Herrmann KM, Weaver LM. 1999. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol. 50:473–503.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lane JF, Koch WT, Leeds NS, Gorin G. 1952. On the toxin of Illicium Anisatum. I. The isolation and characterization of a convulsant principle: anisatin1. J Am Chem Soc. 74:3211–3215.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Preprint arXiv. 13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Morris AB, Bell CD, Clayton JW, Judd WS, Soltis DE, Soltis PS. 2007. Phylogeny and divergence time estimation in Illicium with implications for new world biogeography. Syst Bot. 32:236–249.
- Park J, Yongsung K, Kanghyup L. The complete chloroplast genome of Korean mock strawberry, Duchesnea chrysantha (Zoll. & Moritzi) Miq.(Rosoideae). Mitochondrial DNA Part B. 4:864–865.
- Parthasarathy VA, Chempakam B, Zachariah TJ. 2008. Chemistry of spices. CABI.319-330
- Shen Y, Van Beek TA, Claassen FW, Zuilhof H, Chen B, Nielen MW. 2012. Rapid control of Chinese star anise fruits and teas for neurotoxic anisatin by direct analysis in real time high-resolution mass spectrometry. J Chromatogr A. 1259:179–186.
- Smith AC. 1947. Families Illiciaceae and Schisandraceae. Sargentia. 7:1-224
- Yamada K, Takada S, Nakamura S, Hirata Y. 1968. The structures of anisatin and neoanisatin: toxic sesquiterpenes from Illicium anisatum L. Tetrahedron. 24:199–229.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12:S2.
