Abstract
Pleione formosana is an Endangered terrestrial orchid with significant ornamental values. Here, we report the first complete chloroplast genome of P. formosana. The circular genome was 159,843 bp in length and consisted of a pair of inverted repeats (IR 26,879 bp), which were separated by a large single copy region (LSC 87,392 bp) and a small single copy region (SSC 18,693 bp). It contained 135 genes (114 unique), including 87 protein-coding genes, 38 tRNAs, and 8 rRNAs. The maximum likelihood phylogenetic analysis indicated that P. formosana and P. bulbocodioides cluster together are closely related to the genera Bletilla.
Pleione formosana, Hayata (Citation1911), is a terrestrial orchid, which grows on humus-covered soil or rocks in montane forests and at forest margins, between 600 and 1500 m in southeastern China (1500 and 2500 m in Taiwan island) (Chen et al. Citation2009). Pleione formosana is a vulnerable species in the Red List (IUCN Citation2018) and one of the most important parents for a succession of hybrids. Therefore, it is urgent to establish a strategy for the conservation of this important plant. In addition, Pleione formosana belongs to P. bulbocodioides complex (Cribb and Butterfield Citation1999), which has been thought to be the source of persistent confusion and ambiguity in the taxonomy of Pleione (Gravendeel et al. Citation2004). In recent years, comparative analysis of the complete chloroplast (cp) genome of different close species has provided a promising method for the study of phylogeny, population dynamics, and species evolution (Shaw et al. Citation2014). Thus, we aimed to assemble and characterize P. formosana cp genomes to provide a better understanding of the evolution and genetics of genera Pleione.
Total genomic DNA was extracted from fresh leaves (Voucher specimen: 23°32′25.19″N 120°47′57.71″E, AL01A12, FAFU) using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced by the Illumina Hiseq 2000 sequencing platform (Illumina, CA, USA) at Novogene (Beijing, China). Raw reads were filtered by NGS QC Toolkit (Patel and Jain Citation2012). The clean reads were firstly aligned to P. bulbocodioides (Genbank Accession No. KY849819) (Shi et al. Citation2017). Filtered reads were then assembled into contigs in the software CLC Genomics Workbench v8.0 (CLC Bio, Aarhus, Denmark). After assembled, the obtained scaffolds and contigs were assembled into cp genome by Geneious 11.1.15 (Kearse et al. Citation2012) using the algorithm MUMmer. The genome was automatically annotated using DOGMA (Wyman et al. Citation2004), then adjusted by Geneious version 11.1.15 (Kearse et al. Citation2012) and submitted to GenBank with accession number MK361027. The cp genome sequence of P. formosana is 159,843 bp in length, containing a large single copy (LSC) region of 87,392 bp and a small single copy (SSC) region of 18,693 bp, and two inverted repeat (IR) regions of 26,879 bp. The cp genome encoded 135 genes, of which 114 were unique genes (87 protein-coding genes, 38 tRNAs, and 8 rRNAs). Overall, GC content of the whole genome is 37.2%, while the corresponding values of the LSC, SSC, and IR regions are 35.0, 30.3, and 43.1%, respectively.
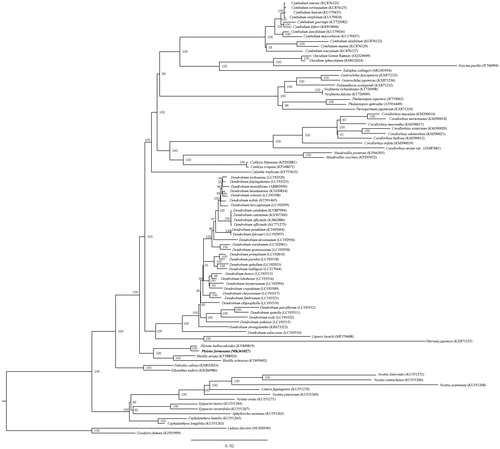
To further investigate its phylogenetic position, 88 complete cp genomes of Epidendroideae and two species of Orchidoideae were aligned using HomBlocks pipeline (Bi et al. Citation2018). RAxML-HPC Black-Box version 8.1.24 (Stamatakis et al. Citation2008) was used to construct a maximum likelihood tree with Ludisia discolor and Goodyera fumata as outgroup. The branch support was computed with 1000 bootstrap replicates. The ML tree analysis indicated that P. formosana and P. bulbocodioides cluster together and are closely related to the genera Bletilla with 100% bootstrap support ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110:18-22.
- Chen X, Cribb PJ, Gale SW. 2009. Pleione. In: Wu ZY, Raven PH, and Hong DY, editors. Flora of China 25 (Orchidaceae). Beijing: Science Press; St. Louis (MO): Missouri Botanical Garden Press; p. 325–333.
- Cribb P, Butterfield I. 1999. The genus Pleione. 2nd ed. Kew: Royal Botanic Gardens; p. 110.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gravendeel B, Eurlings MCM, Berg CVD, Cribb PJ. 2004. Phylogeny of Pleione (Orchidaceae) and parentage analysis of its wild hybrids based on plastid and nuclear ribosomal ITS sequences and morphological data. Syst Bot. 29:50–63.
- Hayata B. 1911. Icones of the plants of Formosa and materials for a flora of the island, based on a study of the collections of the Botanical survey of the government of Formosa. Taihoku: Bureau of Productive Industry, Government of Formosa; p. 64.
- IUCN. 2018. The IUCN red list of Threatened species. Version. 2018-2. [accessed 2018 Dec 20]. www.iucnredlist.org.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Shaw J, Shafer HL, Leonard OR, Kovach MJ, Schorr M, Morris AB. 2014. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am J Bot. 101:1987–2004.
- Shi YC, Yang LF, Yang ZY, Ji YH. 2017. The complete chloroplast genome of Pleione bulbocodioides (Orchidaceae). Conservation Genet Resour. 1:21–25.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.