Abstract
In this study, we determined the complete plastome sequence of Gonocaryum lobbianum (Miers) Kurz (Cardiopteridaceae) (NCBI acc. no. MK390345). This is the first reported complete plastome sequence from the family Cardiopteridaceae of the order Aquifoliales. The complete plastome of G. lobbianum is 158,905 bp in length and consists of a large single copy of 87,705 bp and a small single copy of 19,346 bp, separated by two inverted repeats of 25,927 bp. The plastome contains 112 genes, of which 78 are protein-coding genes, 30 are tRNA genes, and four are rRNA genes. The rpoA gene was pseudogenized because of deletion and following frame-shift mutations. We also confirmed the pseudonization of rpoA gene using PCR and conventional Sanger sequencing methods. Sixteen genes contain one intron and two genes have two introns. The average A-T content of the plastome is 62.7%. A total of 73 simple sequence repeat loci were identified within the genome. Phylogenetic analysis revealed that G. lobbianum is a sister group of Aquifoliaceae-Helwingiaceae with 100% bootstrap value. The Aquifoliales clade is a sister group to the Apiales-Asterales-Dipsacales clade with 100% bootstrap support.
Gonocaryum lobbianum (Miers) Kurz is distributed to south China and southeastern Asia. It belongs to the family Cardiopteridaceae in the Aquifoliales (APG IV Citation2016). The family Cardiopteridaceae contains five genera and about 43 species (Christenhusz and Byng Citation2016). The genus Gonocaryum consists of approximately 10 species. To date, there have been no published plastome sequences for plants in Cardiopteridaceae. We report the first complete plastome sequence of G. lobbianum from the family Cardiopteridaceae. It is expected to become a standard reference sequence of plastome to understanding evolution and phylogenetic relationship of this family.
The leaves of G. lobbianum used in this study were collected in Laos. A voucher specimen was deposited in the Korea University Herbarium (KUS acc. no. TL2008-1633). The collected leaves are dried in silica gel and ground into powder in liquid nitrogen. Total DNAs were extracted using the G-spin™ IIp for Plant Genomic DNA Extraction Kit (iNtRON Biotechnology). The genomic DNAs are deposited in the Plant DNA Bank in Korea (PDBK acc. no. TL2008-1633). The complete plastome sequence was generated using an Illumina MiSeq system (Illumina, Inc., San Diego, CA). An average coverage of sequence was 240.7 times of the G. lobbianum plastome. Annotations were performed using the National Center for Biotechnology Information (NCBI) BLAST, and tRNAscan-SE programs (Lowe and Eddy Citation1997).
The gene order and structure of the G. lobbianum plastome are similar to those of a typical angiosperm (Shinozaki et al. Citation1986; Kim and Lee Citation2004; Yi and Kim Citation2012). The complete plastome is 158,905 bp in length and consists of a LSC of 87,705 bp and a small single copy (SSC) of 19,346 bp, separated by two inverted repeats (IR) of 25,927 bp. The plastome comprises 112 unique genes (78 protein-coding genes, 30 tRNA genes, and four rRNA genes). The average A-T content of the plastome is 62.7%. The A-T contents in the LSC, SSC, and IR regions are 64.0%, 67.6%, and 57.0%, respectively. Sixteen genes contain intron and two genes, ycf3 and clpP, have two introns. The rpoA gene was pseudogenized. A total of 73 simple sequence repeat (SSR) loci are scattered among the plastome. Among these, 62, 7, and 4 are mono-SSR, di-SSR, and tri-SSR loci, respectively.
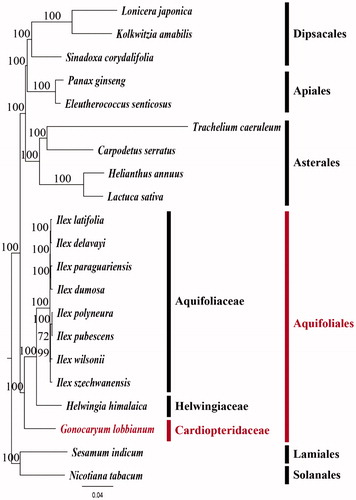
To validate the phylogenetic relationships of G. lobbianum among campanulids, we constructed a maximum likelihood (ML) tree (). Phylogenetic analysis was performed on a data set that included the 79 protein-coding genes and four rRNA genes from 28 taxa using RAxML v. 8.1.17 (Stamatakis et al. Citation2008). The 83 gene sequences (81,856 bp) were aligned with MUSCLE in Geneious v. 11.1.5 (Biomatters Ltd.; Kearse et al. Citation2012). The family Cardiopteridaceae, represented by G. lobbianum, is a sister group to the Hewingiaceae-Aquifoliaceae clade with a 100% bootstrap support. In previous phylogenetic study, Aquifoliales consist of two clades, Aquifoliaceae-Phyllonomiaceae-Phyllonomaceae and Cardiopteridaceae-Stemonuraceae (Tank and Donoghue Citation2010). Therefore, our new plastomes sequences of G. lobbianum represents a Cardiopteridaceae-Stemonuraceae clade. In broader phylogenetic scale, the order Aquifoliales are positioned as sister to the Apiales-Asterales-Dipsacales clade with a 100% bootstrap support. It is consistent with the previous phylogenetic studies, which Aquifoliales is a sister group of the other campanulids (Tank and Donoghue Citation2010; Ruhfel et al. Citation2014).
Figure 1. Maximum Likelihood (ML) tree based on 79 protein-coding and four rRNA genes from 21 plstomes as determined by RAxML(-ln L = 294507.154497). The numbers at each node indicate the ML bootstrap values. Genbank accession numbers of taxa are shown below, Calpodetus serratus (NC_036084), Eleutherococcus senticosus (NC_016430), Gonocaryum lobbianum (MK390345), Helianthus annuus (NC_007977), Helwingia himalaica (NC_031370), Ilex elavayi (KX426470), Ilex dumosa (KP016927), Ilex latifolia (KX426465), Ilex paraguariensis (KP016928), Ilex polyneura (KX426468), Ilex pubescens (KX426467), Ilex szechwanensis (KX426466), Ilex wilsonii (KX426471), Kolkwitzia amabilis (NC_029874), Lactuca sativa (NC_007578), Lonicera japonica (NC_026839), Nicotiana tabacum (NC_001879), Panax ginseng (NC_006290), Sesamum indicum (NC_016433), Sinadoxa corydalifolia (NC_032040), and Trachelium caeruleum (NC_010442).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Christenhusz MJ, Byng JW. 2016. The number of known plants species in the world and its annual increase. Phytotaxa. 261:201–217.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim K-J, Lee H-L. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. 2014. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol. 14:23.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. Embo J. 5:2043–2049.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tank DC, Donoghue MJ. 2010. Phylogeny and phylogenetic nomenclature of the Campanulidae based on an expanded sample of genes and taxa. Syst Bot. 35:425–441.
- Yi D-K, Kim K-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872.
