Abstract
In this study, we determined the complete plastome sequence of Bunchosia argentea (Jacq.) DC. (Malpighiaceae) (NCBI acc. no. MK390344). The gene order and the structure of the B. argentea plastome are collinear with those of the typical plastomes of land plants. The complete plastome size is 158,741 bp in length and consists of a large single-copy region of 88,345 bp and a small single-copy region of 16,816 bp, which are separated by a pair of 26,790 bp-long inverted repeat regions. The overall A-T content of the plastome sequence is 63.5%. The plastome contains 110 genes, of which 76 are protein-coding genes, 30 are tRNA genes, and four are rRNA genes. The rpl32 was lost and two genes (infA and rps7) were pseudogenized. Sixteen genes contain one intron and two genes have two introns. A total of 87 simple sequence repeat loci were identified within the genome. Phylogenetic analysis revealed that B. argentea and two Byrsonima species formed a clade with 100% bootstrap support.
Bunchosia argentea (Jacq.) DC., commonly referred to as peanut butter fruit, originated in South America. It belongs to the family Malpighiaceae in the order Malpighiales (APG IV Citation2016).
The leaves of B. argentea used in this study were collected from a farm in Malaysia. A voucher specimen was deposited in the Korea University Herbarium (KUS acc. no. TM2017-1523). The silica gel dried leaves were ground into powder in liquid nitrogen and total DNAs were extracted using the G-spin™ IIp for Plant Genomic DNA Extraction Kit (iNtRON Biotechnology). The genomic DNAs are deposited in the Plant DNA Bank in Korea (PDBK acc. no. 2017-1523). The complete plastome sequence was generated using an Illumina MiSeq platform (Illumina Inc., San Diego, CA). Annotations were performed using the find annotation by Geneious 11.1.5 (Biomatters Ltd.; Kearse et al. Citation2012), National Center for Biotechnology Information (NCBI) BLAST and tRNAscan-SE programs (Lowe and Eddy Citation1997).
The gene order and the structure of the B. argentea plastome are similar to those of typical angiosperms (Shinozaki et al. Citation1986; Kim and Lee Citation2004; Yi and Kim Citation2012). The complete plastome is 158,741 bp in length and consists of a large single-copy (LSC) region of 88,345 bp and a small single-copy (SSC) region of 16,816 bp, which are separated by two inverted repeats (IR) of 26,790 bp. The plastome comprises 110 unique genes (76 protein-coding genes, 30 tRNA genes, and four rRNA genes). The rpl32 gene was lost. The infA and rps7 were pseudogene. These results were reported from Malpighiales and Oxalidales (Tangphatsornruang et al. Citation2011; Huang et al. Citation2014; Bardon et al. Citation2016; Jo et al. Citation2016, Citation2017). Seven protein-coding, seven tRNA, and four rRNA genes are duplicated in the IR regions. The average A-T content of the plastome is 63.5%, whereas that in the LSC, SSC, and IR regions is 65.9%, 69.8%, and 57.7%, respectively. Sixteen genes contain one intron and two genes, ycf3 and clpP, have two introns. A total of 74 simple sequence repeat (SSR) loci are distributed throughout the plastome. Among these, 62, 9, and 3 are mono-SSR, di-SSR, and tri-SSR loci, respectively. Some of these loci will be useful in identifying cultivars of B. argentea.
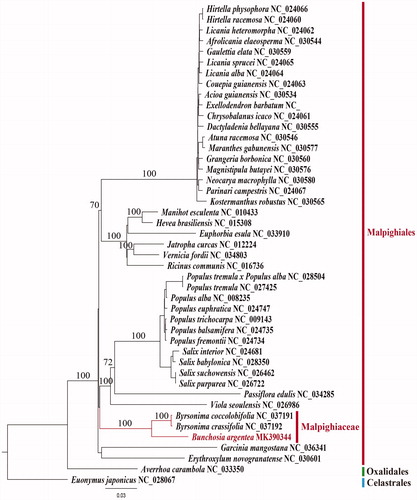
For the phylogenetic analysis, we selected and downloaded 44 complete plastome sequences from the NCBI database. The plastome sequences were selected from a COM (Celastrales-Oxalidales-Malpighiales) clade. The aligned 76 protein-coding and four rRNA gene sequences using MUSCLE program in Geneious v. 11.1.5 (Biomatters Ltd.; Kearse et al. Citation2012) were 78,665 bp in length. The RAxML tree (Stamatakis et al. Citation2008) shows that B. argentea forms a clade with two Byrsonima species with a 100% bootstrap support (). Malpighiaceae forms a clade with Salicaceae-Passifloraceae-Violaceae members. However, the clade has a lower resolution with less than 50% bootstrap support. The close relationship of Malpighiaceae to Elatinaceae was suggested by a previous study (Davis and Anderson Citation2010; Xi et al. Citation2012), but the complete plastome sequences is not available from Elatinaceae. Malphigiaceae consist of two subfamilies, Byrsonimoideae and Malpighioideae. Previously, two plastome sequences were available from the subfamily Byrsonimoideae. Our new addition of plastome sequence from the subfamily Malpighioideae, B. argentea, provide a useful resource for the phylogenetic understanding of Malpighiaceae.
Disclosure statement
The authors report no conflicts of interest and are independently responsible for the content and writing of the paper.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181:1–20.
- Bardon L, Sothers C, Prance GT, Malé P-JG, Xi Z, Davis CC, Murienne J, García-Villacorta R, Coissac E, Lavergne S, Chave J. 2016. Unraveling the biogeographical history of Chrysobalanaceae from plastid genomes. Am J Bot. 103:1089–1102.
- Davis CC, Anderson WR. 2010. A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Am J Bot. 97:2031–2048.
- Huang DI, Hefer CA, Kolosova N, Douglas CJ, Cronk QC. 2014. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae). New Phytol. 204:693–703.
- Jo S, Kim H-W, Kim Y-K, Cheon S-H, Kim K-J. 2016. Complete plastome sequence of Averrhoa carambola L.(Oxalidaceae). Mitochondr DNA B Resour. 1:609–611.
- Jo S, Kim H-W, Kim Y-K, Sohn J-Y, Cheon S-H, Kim K-J. 2017. The complete plastome of tropical fruit Garcinia mangostana (Clusiaceae). Mitochondr DNA B Resour. 2:722–724.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim K-J, Lee H-L. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 11:247–261.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. 1986. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. Embo J. 5:2043–2049.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57:758–771.
- Tangphatsornruang S, Uthaipaisanwong P, Sangsrakru D, Chanprasert J, Yoocha T, Jomchai N, Tragoonrung S. 2011. Characterization of the complete chloroplast genome of Hevea brasiliensis reveals genome rearrangement, RNA editing sites and phylogenetic relationships. Gene. 475:104–112.
- Xi Z, Ruhfel BR, Schaefer H, Amorim AM, Sugumaran M, Wurdack KJ, Endress PK, Matthews ML, Stevens PF, Mathews S, Davis CC. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc Natl Acad Sci USA. 109:17519–17524.
- Yi D-K, Kim K-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS One. 7:e35872.