Abstract
Anopheles funestus is a major malaria vector in Africa. In this study, its complete mitochondrial genome was assembled from high-throughput sequencing reads. The circular genome is 15,403 bp long with an A + T-biased base composition and harbors 37 genes (including 13 protein-coding genes/PCGs, 22 tRNAs, and two rRNAs) and one control region. The PCGs are initiated with ATN, GTG, TTG, or CGA codons and are terminated with either TAA or TA/T codons. Phylogenetic analysis added support to the current taxonomic framework of the genus Anopheles and suggested that A. funestus is closely related to A. culicifacies and A. minimus.
Malaria remains a major public health issue in Africa, with Anopheles funestus and Anopheles gambiae being the predominant vectors (Djouaka et al. Citation2016). Anopheles funestus is regarded as being among the first species that have adapted to human hosts (Charlwood et al. Citation1995) and far outstrips A. gambiae in its ability to transmit Plasmodium falciparum in some cases (Coetzee and Fontenille Citation2004). This mosquito is morphologically identical at all life stages to three other members of the funestus subgroup (A. aruni, A. parensis, and A. vaneedeni) of the funestus group (series Myzomyia within subgenus Cellia) (Coetzee and Fontenille Citation2004). To facilitate the accurate identification of A. funestus, we assembled its mitochondrial genome in this study. The annotated sequence has been deposited into GenBank under the accession number MF775371.
Totally, 91.4 M 125-bp-long raw reads (SRA accession: ERR1358729) were retrieved from a previously published study (Barnes et al. Citation2017). These reads were originally produced from individual females collected from Chikwawa, Malawi (16°3’S, 34°50’E; NCBI BioSample: SAMEA3928332). Following the trimming with Trimmomatic v0.35 (Bolger et al. Citation2014), they were employed to assemble the mitochondrial genome using the ARC program (Hunter et al. Citation2015), with that of A. gambiae (NC_002084) (Beard et al. Citation1993) as the seed reference. The genome was annotated using the MITOS web server (Bernt et al. Citation2013), followed by delicate adjustment as described by Cameron (Citation2014).
The circular genome of A. funestus is 15,403 bp in size and has a highly asymmetric base composition (40.3%A, 12.6%C, 9.1%G & 38.0%T; light strand). It encodes 13 protein-coding genes/PCGs, 22 tRNAs, and two rRNAs and harbors a non-coding control region. Its genomic organization is identical to those of its congeners (Peng et al. Citation2016). Most genes are located on the heavy strand except for four PCGs, nine tRNAs, and the two rRNAs.
All PCGs are initiated with ATN codons except for COX1 (CGA), ND1 (TTG), and ND5 (GTG) and are terminated with either TAA (ATP6, ATP8, ND1, ND4L, and ND6) or TA/T (the remaining eight PCGs) codon. The 22 tRNAs range in size from 64 to 72 bp with a total length of 1470 bp. The two rRNAs are separated by tRNA-Val and are 791 bp (12S rRNA) and 1363 bp (16S rRNA) in length, respectively. The control region is 570 bp long with a remarkably high A + T content (94.0%) and is located between 12S rRNA and tRNA-Ile. Seven intergenic spacer regions are present across the genome, ranging in size from 1 to 34 bp with a total length of 76 bp. There are 10 intergenic overlapping regions ranging in size from 1 and 7 bp with a total length of 26 bp.
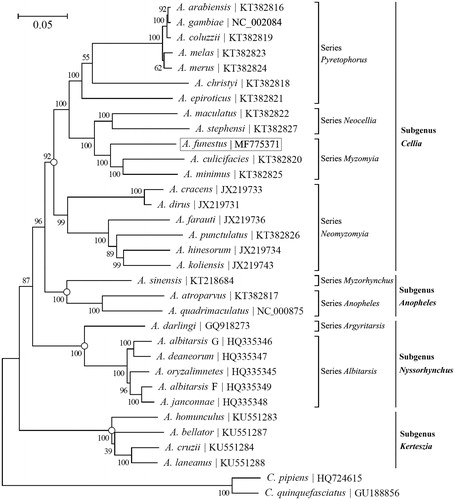
To investigate its taxonomic placement, a maximum-likelihood (ML) phylogenetic tree was reconstructed using the concatenated sequences of all 13 PCGs with MEGA7 (Kumar et al. Citation2016) (). The phylogeny recovered here corroborates the validity of the series- and subgenus-level taxonomy of Anopheles mosquitoes and suggests that A. funestus is phylogenetically close to A. culicifacies and A. minimus.
Figure 1. Phylogeny of 31 Anopheles mosquitoes based on the maximum likelihood (ML) analysis of the concatenated sequence of 13 PCGs. The ‘GTR + G+I’ substitution model was employed as suggested by MEGA7 (Kumar et al. Citation2016). The bootstrap values are based on 500 resamplings. The tree was rooted with Culex pipiens and Culex quinquefasciatus. Codon positions included are 1st + 2nd + 3rd.
Acknowledgements
The authors thank Dr. Kayla G. Barnes and her colleagues for generating the genomic data used in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barnes KG, Weedall GD, Ndula M, Irving H, Mzihalowa T, Hemingway J, Wondji CS. 2017. Genomic footprints of selective sweeps from metabolic resistance to pyrethroids in African malaria vectors are driven by scale up of insecticide-based vector control. PLoS Genet. 13:e1006539
- Beard CB, Hamm DM, Collins FH. 1993. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 2:103–124.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Cameron SL. 2014. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst Entomol. 39:400–411.
- Charlwood JD, Smith T, Kihonda J, Heiz B, Billingsley PF, Takken W. 1995. Density independent feeding success of malaria vectors (Diptera: Culicidae) in Tanzania. BER. 85:29–35.
- Coetzee M, Fontenille D. 2004. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem Mol Biol. 34:599–605.
- Djouaka R, Akoton R, Tchigossou GM, Atoyebi SM, Irving H, Kusimo MO, Djegbe I, Riveron JM, Tossou E, Yessoufou A, et al. 2016. Mapping the distribution of Anopheles funestus across Benin highlights a sharp contrast of susceptibility to insecticides and infection rate to Plasmodium between southern and northern populations. Wellcome Open Res. 1:28.
- Hunter SS, Lyon RT, Sarver BAJ, Hardwick K, Forney LJ, Settles ML. 2015. Assembly by Reduced Complexity (ARC): a hybrid approach for targeted assembly of homologous sequences. bioRxiv. https://doi.org/10.1101/014662.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Peng X-Y, Zhou P, Duan X-Y, Qian Z-Q. 2016. The mitochondrial genomes of twelve Anopheles mosquitoes (Diptera: Culicidae) and their phylogenetic implications. Conservation Genet Resour. 8:387–390.
