Abstract
Rochia nilotica is a common marine gastropod species in family Tegulidae. At present, less molecular information of genus Rochia was reported. In the present study, we first determined the complete mitochondrial genome of R. nilotica. The mitochondrial sequence was 16,966 bp in length and encoded for 35 metazoan mitochondrial genes, including 13 protein-coding genes, 20 transfer RNAs, and two rRNAs. The overall nucleotide composition is 32.2% A, 17.3% C, 17.8% G, and 32.7% T. Thirteen protein-coding genes started with ATG codons, and 10 protein-coding genes used termination codon of TAA, whereas the remaining three had a termination codon of TAG. Three overlaps between adjacent genes were found in the mitochondrial gene. The phylogenetic relationships of 17 mitochondrial sequences based on the concatenated nucleotide sequences of 13 protein-coding genes showed that R. nilotica was closely related to Tectus pyramis.
Individuals of Rochia nilotica were collected from the coastal waters of Quanfu Island, South China Sea and preserved in 95% ethanol in the collection of Ocean University of China. Total genomic DNA was extracted from the foot muscle, using cetyltrime thylammonium bromide (CTAB) extraction method (Schulenburg et al. Citation2001). Total genomic DNA was sequenced on an Illumina MiSeq sequencer, using a PE150 protocol. Short-read DNA sequences were assembled using CLC Genomics Workbench 11 (CLC Bio, Aarhus, Denmark). Mitochondrial protein-coding genes (PCGs) were annotated by identifying their open reading frames with ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) and blasted using the invertebrate mitochondrial genetic code. The transfer RNA (tRNA) genes were identified using MITOS web servers (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). The ribosomal (rrnL and rrnS) genes were identified by their similarity to gene sequence of Tectus pyramis. The nucleotide compositions were computed using MEGA 6 (Tamura et al. Citation2013).
The mitochondrial sequence (GenBank accession MK284240) was 16,966 bp in length and encoded for 35 metazoan mitochondrial genes, including 13 PCGs, 20 transfer RNAs, and two rRNAs. Thirteen PCGs started with ATG codons, and 10 PCGs used termination codon of TAA, whereas the remaining three had a termination codon of TAG. The overall nucleotide composition was 32.2% A, 17.3% C, 17.8% G, and 32.7% T. Three overlaps between adjacent genes were found in the mitochondrial gene, ranging from 5bp to 151bp in length. As expected, R. nilotica showed a highly conserved genome structure of ancestral gastropods (Lee et al. Citation2016).
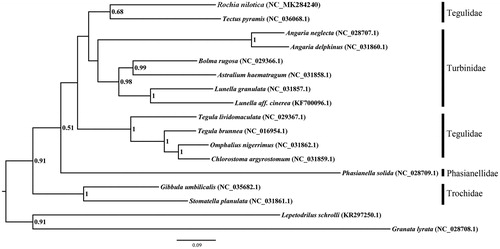
Maximum Likelihood (ML) method was used to perform the phylogenetic analysis, based on nucleotide sequences of 13 concentrated PCGs of 17 available mitochondrial genomes, and Lepetodrilus schrolli and Granata lyrata were set as out group (). According to our study, R. nilotica was clustered with T. pyramis, and the clade (Rochia + Tectus) had a close relationship with Turbinidae than with other Tegulidae, indicating Tectus and Rochia need to be assigned to a new family (Williams Citation2012; Lee et al. Citation2016; Uribe et al. Citation2017).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Lee H, Samadi S, Puillandre N, Tsai MH, Dai CF, Chen WJ. 2016. Eight new mitogenomes for exploring the phylogeny and classification of Vetigastropoda. J. Molluscan Stud. 82:534–541.
- Schulenburg JHGV, Hancock JM, Pagnamenta A, Sloggett JJ, Majerus MEN, Hurst GDD. 2001. Extreme length and length variation in the first ribosomal internal transcribed spacer of ladybird beetles (Coleoptera: Coccinellidae). Mol Biol Evol. 18: 648–660.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Uribe JE, Williams ST, Templado J, Abalde S, Zardoya R. 2017. Denser mitogenomic sampling improves resolution of the phylogeny of the superfamily Trochoidea (Gastropoda: Vetigastropoda). J. Molluscan Stud. 83:111–118.
- Williams ST. 2012. Advances in molecular systematics of the vetigastropod superfamily Trochoidea. Zool Scr. 41:571–595.