Abstract
Coffea arabica is major cultivated species of coffee. We selected cold hardiness of C. arabica (named as CH3) based on selection of coffee seeds in Jeju Island, Korea. Here, we presented complete chloroplast genome of cold resistance C. arabica which is 155,192 bp long and has four subregions: 85,163 bp of large single copy (LSC) and 18,137 bp of small single copy (SSC) regions are separated by 25,946 bp of inverted repeat (IR) regions including 131 genes (86 protein-coding genes, eight rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 37.4% and those in the LSC, SSC, and IR regions are 35.3%, 31.3%, and 43.0%, respectively. Three non-synonymous single nucleotide polymorphisms and three insertions and deletions are found, indicating that further analysis will be required to understand genetic elements of cold hardiness of CH3.
Coffea arabica L., cultivated for producing coffee products occupying 70% of world coffee production (Lashermes et al. Citation1999; O'brien and Kinnaird Citation2003), is originated from forests of southwestern highlands in Ethiopia (Barre et al. Citation1998). Coffea arabica was formed by hybridization of Coffea canephora and Coffea eugenioides (Lashermes et al. Citation1999). C. arabica has been distributed by human for coffee production, forming ‘Bean belt’ covering from S20° to N20° (Bentley and Baker Citation2000). This limited range is determined by the fact that C. arabica leaves cannot be recovered once it is exposed under 5 °C (Willson Citation1999). Some pioneers, who aim for commercial coffee production in Korea (N33° to N38°), have tried finding cold hardiness individuals by selecting seeds in Jeju island, South Korea. More than 10 years ago, Mr. Nho, one of the authors, successfully selected coffee tree which can survive at −2 °C. It is healthy to produce 1 to 2 kg coffee beans per year entitled coffee beans cultivated in Korea.
To understand genetic elements of cold resistant coffee tree (named as CH3; YS. Kim, IB-00579 in InfoBoss Cyber Herbarium (IN)), its total DNA was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea, and de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011). All assembled sequences were confirmed using BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for chloroplast genome annotation based on C. arabica chloroplast genome (NC_008535; Samson et al. Citation2007).
The CH3 chloroplast genome (Genbank accession is MK342634) is 155,192 bp and has four subregions: 85,163 bp of large single copy (LSC) and 18,137 bp of small single copy (SSC) regions are separated by 25,946 bp of inverted repeat (IR). It contains 131 genes (86 protein-coding genes, eight rRNAs, and 37 tRNAs); 19 genes (eight protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions. The overall GC content is 37.4% and those in the LSC, SSC, and IR regions are 35.3%, 31.3%, and 43.0%, respectively.
Based on alignment of published C. arabica chloroplast genome (NC_008535), three single nucleotide polymorphisms and three insertion and deletions were identified. All SNPs are non-synonymous in ycf1 and ndhF and all INDELs are in intergenic space.
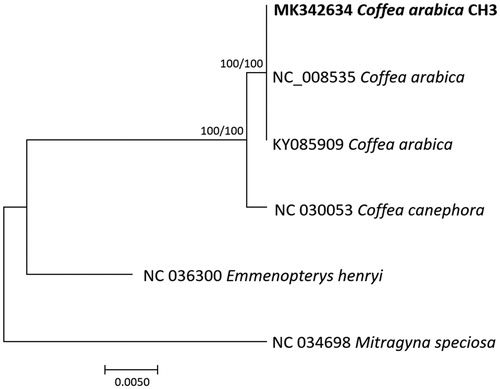
Four Coffea and two chloroplast genomes of Rubiaceae as outgroup species were used for constructing phylogenic trees. Whole chloroplast genome sequences were aligned by MAFFT 7.388 (Katoh and Standley Citation2013) for constructing neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) trees using MEGA X (Kumar et al. Citation2018). Phylogenetic trees show that three C. arabica are clustered with C. canephora (Wu et al. Citation2017), one of parental species of C. arabica (; Lashermes et al. Citation1999). Without CH3-specific sequence variations, genetic elements linked to cold hardiness will be identified through further analyses of its mitochondrial or whole genomes.
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1000) phylogenetic trees of four Coffea and two outgroup complete chloroplast genomes: three Coffea arabica (MK342634, in this study, NC_008535 and KY085909), Coffea canephora (NC_030053), Mitragyna speciosa (NC_034698), and Emmenopterys henryi (NC_036300). Phylogenetic tree was drwon based on neighbor joining tree. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic tree, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barre P, Layssac M, D’Hont A, Louarn J, Charrier A, Hamon S, Noirot M. 1998. Relationship between parental chromosomic contribution and nuclear DNA content in the coffee interspecific hybrid C. pseudozanguebariae× C. liberica var ‘dewevrei’. Theoretical and Applied Genetics. 96:301–305.
- Bentley JW, Baker PS. 2000. The Colombian Coffee Growers' Federation: organised, successful smallholder farmers for 70 years. Bogotá (Colombia): ODI.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549.
- Lashermes P, Combes M-C, Robert J, Trouslot P, D'Hont A, Anthony F, Charrier A. 1999. Molecular characterisation and origin of the Coffea arabica L. genome. Mol General Genet MGG. 261:259–266.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- O'brien TG, Kinnaird MF. 2003. Caffeine and conservation. Am Assoc Adv Sci. 300:587.
- Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. 2007. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol J. 5:339–353.
- Willson K. 1999. Coffee, cocoa ant tea. Wallingford: CAB International.
- Wu D, Bi C, Wang X, Xu Y, Ye Q, Ye N. 2017. The complete chloroplast genome sequence of an economic plant Coffea canephora. Mitochondr DNA B. 2:483–485.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinform. 12:S2.
